













| General Information: |
|
| Name(s) found: |
RAD23 /
YEL037C
[SGD]
|
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 398 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleotide-excision repair factor 2 complex
[IDA mitochondrion [IDA repairosome [IDA proteasome complex [IMP |
| Biological Process: |
nucleotide-excision repair, DNA damage recognition
[TAS negative regulation of protein catabolic process [TAS ER-associated protein catabolic process [IMP |
| Molecular Function: |
damaged DNA binding
[IDA protein binding, bridging [IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVSLTFKNFK KEKVPLDLEP SNTILETKTK LAQSISCEES QIKLIYSGKV LQDSKTVSEC 60
61 GLKDGDQVVF MVSQKKSTKT KVTEPPIAPE SATTPGRENS TEASPSTDAS AAPAATAPEG 120
121 SQPQEEQTAT TERTESASTP GFVVGTERNE TIERIMEMGY QREEVERALR AAFNNPDRAV 180
181 EYLLMGIPEN LRQPEPQQQT AAAAEQPSTA ATTAEQPAED DLFAQAAQGG NASSGALGTT 240
241 GGATDAAQGG PPGSIGLTVE DLLSLRQVVS GNPEALAPLL ENISARYPQL REHIMANPEV 300
301 FVSMLLEAVG DNMQDVMEGA DDMVEGEDIE VTGEAAAAGL GQGEGEGSFQ VDYTPEDDQA 360
361 ISRLCELGFE RDLVIQVYFA CDKNEEAAAN ILFSDHAD |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
PNG1 RAD23 RAD4 |
| View Details | (MIPS) Mewes HW, et al. (2004) |

|
RAD23 RAD4 |
| View Details | Krogan NJ, et al. (2006) |

|
PNG1 RAD23 |
| View Details | Gavin AC, et al. (2002) |

|
DNA2 ECM29 KAP95 MEC1 MPH1 MRPS28 MSH2 MSH6 PGK1 RAD23 RAD34 RAD4 RAD52 RFA1 RFA2 RPN10 RPN12 RPN3 RPN6 RPN8 RPN9 RPT2 RPT3 RPT6 SRO7 SSL1 TFA1 TFB1 TFB4 |
| View Details | Ho Y, et al. (2002) |
|
DSK2 HMF1 NPL4 RAD23 SHP1 TSL1 UBI4 UFD2 YCR001W YDR049W |
| View Details | Qiu et al. (2008) |
|
ABF1 RAD14 RAD16 RAD23 RAD4 RAD7 |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
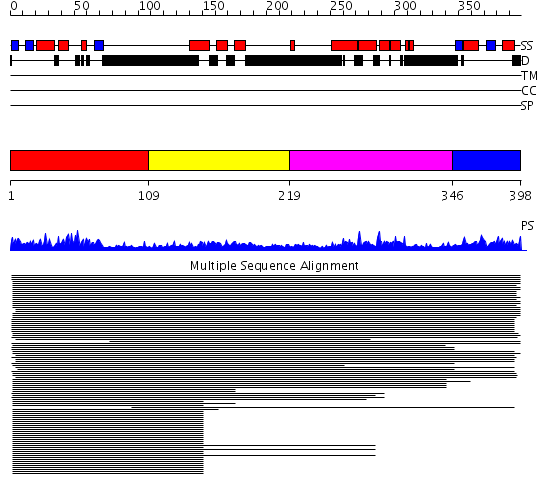
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..108] | 49.519517 | Ubiquitin | |
| 2 | View Details | [109..218] | 102.09691 | DNA repair protein Hhr23a | |
| 3 | View Details | [219..345] | 2.061993 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [346..398] | 74.39794 | DNA repair protein Hhr23a |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)