













| General Information: |
|
| Name(s) found: |
NGG1 /
YDR176W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 702 amino acids |
Gene Ontology: |
|
| Cellular Component: |
SLIK (SAGA-like) complex
[IPI Ada2/Gcn5/Ada3 transcription activator complex [IPI SAGA complex [IDA |
| Biological Process: |
chromatin modification
[TAS response to drug [IMP histone acetylation [TAS |
| Molecular Function: |
transcription cofactor activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPRHGRRGKL PKGEKLPKKE GGDNTPSKLL SSMLKTLDLT FERDIGMLNG KSVRSIPNKK 60
61 TLLELQSQLD SLNEILGTIA RGDQETIEAL RKIRDSKNEK QANDEKQETS NADGQHESST 120
121 ATEETNIIDK GVQSPPKPPP SNEISGTIEN DVESIKQAAD NMAKEEINED KDLQVHRDQP 180
181 REKRPFDSET ENRATENENT QRPDNKKQKI DVDKMENDPT VKNPKSEFVV SQTLPRAAAA 240
241 LGLFNEEGLE STGEDFLKKK YNVASYPTND LKDLLPGELP DMDFSHPKPT NQIQFNTFLA 300
301 FVENFFKDLS DDNLKFLKMK YIIPDSLQFD KTYDPEVNPF IIPKLGPLYT DVWFKDENDK 360
361 NSAYKKPSPY SNDASTILPK KSANELDDNA LESGSISCGP LLSRLLSAVL KDDNDKSELQ 420
421 SSKIIRDGGL PRTGGEDDIQ SFRNNNNDTV DMTLSQENGP SVQTPDNDID EEASFQAKLA 480
481 ENKGSNGGTT STLPQQIGWI TNGINLDYPT FEERLKRELK YVGIYMNLPK DENNPNSDDP 540
541 DWVTGREDDE ISAELRELQG TLKQVTKKNQ KRKAQLIPLV ERQLAWQEYS SILEDLDKQI 600
601 DQAYVKRIRV PKKRKKHHTA ASNNVNTGTT SQIAQQKAAN SSLKSLLDKR QRWINKIGPL 660
661 FDKPEIMKRI PNESVFKDMD QEEDEDEADV FAQNTNKDVE LN |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #04 Alpha Factor Prep2 | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
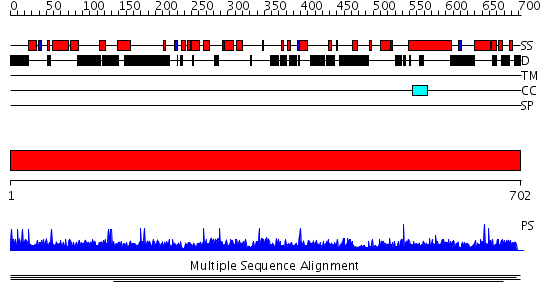
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..702] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)