













| General Information: |
|
| Name(s) found: |
RPS6A /
YPL090C
[SGD]
RPS6B / YBR181C [SGD] |
| Description(s) found:
Found 43 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 236 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA small nucleolar ribonucleoprotein complex [IPI |
| Biological Process: |
translation
[IDA |
| Molecular Function: |
structural constituent of ribosome
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKLNISYPVN GSQKTFEIDD EHRIRVFFDK RIGQEVDGEA VGDEFKGYVF KISGGNDKQG 60
61 FPMKQGVLLP TRIKLLLTKN VSCYRPRRDG ERKRKSVRGA IVGPDLAVLA LVIVKKGEQE 120
121 LEGLTDTTVP KRLGPKRANN IRKFFGLSKE DDVRDFVIRR EVTKGEKTYT KAPKIQRLVT 180
181 PQRLQRKRHQ RALKVRNAQA QREAAAEYAQ LLAKRLSERK AEKAEIRKRR ASSLKA |
The following runs contain data for this protein:
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Hazbun TR, et al. (2003) |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
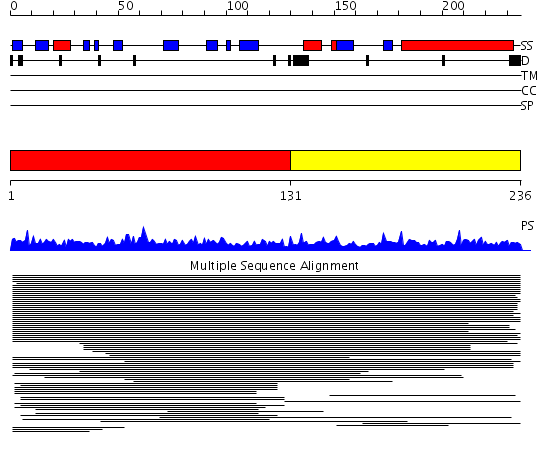
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..130] | 80.853872 | Ribosomal protein S6e No confident structure predictions are available. | |
| 2 | View Details | [131..236] | 10.073995 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)