













| General Information: |
|
| Name(s) found: |
HFD1 /
YMR110C
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 532 amino acids |
Gene Ontology: |
|
| Cellular Component: |
lipid particle
[IDA integral to mitochondrial outer membrane [IDA endosome [IDA mitochondrion [IDA mitochondrial outer membrane [IDA |
| Biological Process: |
cellular aldehyde metabolic process
[IC |
| Molecular Function: |
3-chloroallyl aldehyde dehydrogenase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSNDGSKILN YTPVSKIDEI VEISRNFFFE KQLKLSHENN PRKKDLEFRQ LQLKKLYYAV 60
61 KDHEEELIDA MYKDFHRNKI ESVLNETTKL MNDILHLIEI LPKLIKPRRV SDSSPPFMFG 120
121 KTIVEKISRG SVLIIAPFNF PLLLAFAPLA AALAAGNTIV LKPSELTPHT AVVMENLLTT 180
181 AGFPDGLIQV VQGAIDETTR LLDCGKFDLI FYTGSPRVGS IVAEKAAKSL TPCVLELGGK 240
241 SPTFITENFK ASNIKIALKR IFFGAFGNSG QICVSPDYLL VHKSIYPKVI KECESVLNEF 300
301 YPSFDEQTDF TRMIHEPAYK KAVASINSTN GSKIVPSKIS INSDTEDLCL VPPTIVYNIG 360
361 WDDPLMKQEN FAPVLPIIEY EDLDETINKI IEEHDTPLVQ YIFSDSQTEI NRILTRLRSG 420
421 DCVVGDTVIH VGITDAPFGG IGTSGYGNYG GYYGFNTFSH ERTIFKQPYW NDFTLFMRYP 480
481 PNSAQKEKLV RFAMERKPWF DRNGNNKWGL RQYFSLSAAV ILISTIYAHC SS |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
GIN4 HFD1 HTZ1 KAP114 KCC4 NAP1 PRO3 REI1 RIM11 RPS6A, RPS6B UBC8 VIP1 WTM1 YKR011C |
| View Details | Qiu et al. (2008) |
|
GUT2 HFD1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
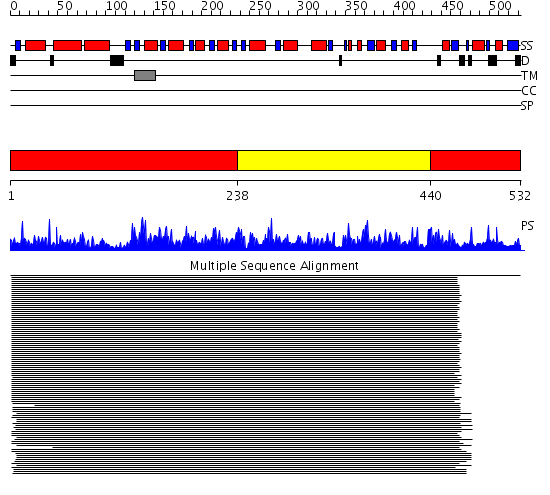
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..237] [440..532] |
1321.54902 | Aldehyde reductase (dehydrogenase), ALDH | |
| 2 | View Details | [238..439] | 1321.54902 | Aldehyde reductase (dehydrogenase), ALDH |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|