













| General Information: |
|
| Name(s) found: |
PUF2 /
YPR042C
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1075 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay
[IGI |
| Molecular Function: |
mRNA binding
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDNKRLYNGN LSNIPEVIDP GITIPIYEED IRNDTRMNTN ARSVRVSDKR GRSSSTSPQK 60
61 IGSYRTRAGR FSDTLTNLLP SISAKLHHSK KSTPVVVVPP TSSTPDSLNS TTYAPRVSSD 120
121 SFTVATPLSL QSTTTRTRTR NNTVSSQITA SSSLTTDVGN ATSANIWSAN AESNTSSSPL 180
181 FDYPLATSYF EPLTRFKSTD NYTLPQTAQL NSFLEKNGNP NIWSSAGNSN TDHLNTPIVN 240
241 RQRSQSQSTT NRVYTDAPYY QQPAQNYQVQ VPPRVPKSTS ISPVILDDVD PASINWITAN 300
301 QKVPLVNQIS ALLPTNTISI SNVFPLQPTQ QHQQNAVNLT STSLATLCSQ YGKVLSARTL 360
361 RGLNMALVEF STVESAICAL EALQGKELSK VGAPSTVSFA RVLPMYEQPL NVNGFNNTPK 420
421 QPLLQEQLNH GVLNYQLQQS LQQPELQQQP TSFNQPNLTY CNPTQNLSHL QLSSNENEPY 480
481 PFPLPPPSLS DSKKDILHTI SSFKLEYDHL ELNHLLQNAL KNKGVSDTNY FGPLPEHNSK 540
541 VPKRKDTFDA PKLRELRKQF DSNSLSTIEM EQLAIVMLDQ LPELSSDYLG NTVIQKLFEN 600
601 SSNIIRDIML RKCNKYLTSM GVHKNGTWVC QKIIKMANTP RQINLVTSGV SDYCTPLFND 660
661 QFGNYVIQGI LKFGFPWNSF IFESVLSHFW TIVQNRYGSR AVRACLEADS IITQCQLLTI 720
721 TSLIIVLSPY LATDTNGTLL ITWLLDTCTL PNKNLILCDK LVNKNLVKLC CHKLGSLTVL 780
781 KILNLRGGEE EALSKNKIIH AIFDGPISSD SILFQILDEG NYGPTFIYKV LTSRILDNSV 840
841 RDEAITKIRQ LILNSNINLQ SRQLLEEVGL SSAGISPKQS SKNHRKQHPQ GFHSPGRARG 900
901 VSVSSVRSSN SRHNSVIQMN NAGPTPALNF NPAPMSEINS YFNNQQVVYS GNQNQNQNGN 960
961 SNGLDELNSQ FDSFRIANGT NLSLPIVNLP NVSNNNNNYN NSGYSSQMNP LSRSVSHNNN1020
1021 NNTNNYNNND NDNNNNNNNN NNNNNNNNNN NNNSNNSNNN NNNDTSLYRY RSYGY |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample: HX13 - trans-SNARE complex forms but fusion is blocked. | Hao Xu, et al. (2010) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #17 Mitotic Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
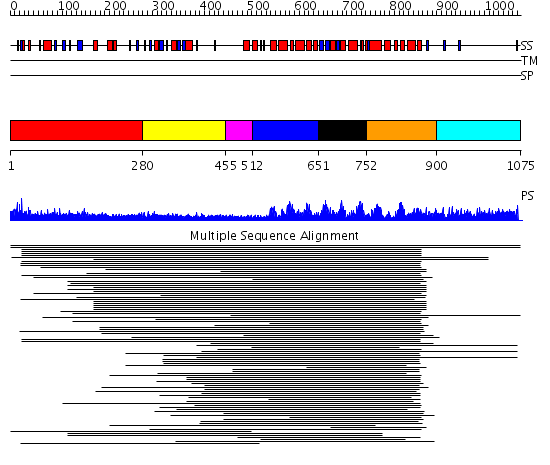
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..279] | 15.438997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [280..454] | 3.040959 | RNA recognition motif. (a.k.a. RRM, RBD, or RNP domain) No confident structure predictions are available. | |
| 3 | View Details | [455..511] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [512..650] | 200.0103 | Pumilio 1 | |
| 5 | View Details | [651..751] | 200.0103 | Pumilio 1 | |
| 6 | View Details | [752..899] | 200.0103 | Pumilio 1 | |
| 7 | View Details | [900..1075] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 4 |
|
|||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 6 |
|
|||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)