













| General Information: |
|
| Name(s) found: |
TPS1 /
YBR126C
[SGD]
|
| Description(s) found:
Found 45 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 495 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA alpha,alpha-trehalose-phosphate synthase complex (UDP-forming) [IDA |
| Biological Process: |
carbohydrate metabolic process
[IDA response to drug [IMP trehalose biosynthetic process [IDA response to stress [IDA |
| Molecular Function: |
alpha,alpha-trehalose-phosphate synthase (UDP-forming) activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTTDNAKAQL TSSSGGNIIV VSNRLPVTIT KNSSTGQYEY AMSSGGLVTA LEGLKKTYTF 60
61 KWFGWPGLEI PDDEKDQVRK DLLEKFNAVP IFLSDEIADL HYNGFSNSIL WPLFHYHPGE 120
121 INFDENAWLA YNEANQTFTN EIAKTMNHND LIWVHDYHLM LVPEMLRVKI HEKQLQNVKV 180
181 GWFLHTPFPS SEIYRILPVR QEILKGVLSC DLVGFHTYDY ARHFLSSVQR VLNVNTLPNG 240
241 VEYQGRFVNV GAFPIGIDVD KFTDGLKKES VQKRIQQLKE TFKGCKIIVG VDRLDYIKGV 300
301 PQKLHAMEVF LNEHPEWRGK VVLVQVAVPS RGDVEEYQYL RSVVNELVGR INGQFGTVEF 360
361 VPIHFMHKSI PFEELISLYA VSDVCLVSST RDGMNLVSYE YIACQEEKKG SLILSEFTGA 420
421 AQSLNGAIIV NPWNTDDLSD AINEALTLPD VKKEVNWEKL YKYISKYTSA FWGENFVHEL 480
481 YSTSSSSTSS SATKN |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi 1 with ha tag from october 2005 | McCusker D, et al (2007) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
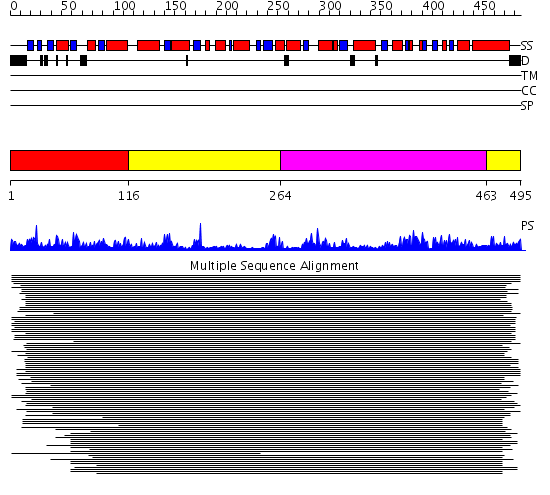
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..115] | 1.41699 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [116..263] [463..495] |
13.21 | UDP-N-acetylglucosamine 2-epimerase | |
| 3 | View Details | [264..462] | 13.21 | UDP-N-acetylglucosamine 2-epimerase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)