













| General Information: |
|
| Name(s) found: |
ADE17 /
YMR120C
[SGD]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 592 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[IDA |
| Biological Process: |
purine nucleotide biosynthetic process
[IDA 'de novo' IMP biosynthetic process [IDA |
| Molecular Function: |
IMP cyclohydrolase activity
[IDA phosphoribosylaminoimidazolecarboxamide formyltransferase activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MANYTKTAIL SVYDKTGLLD LARGLIEKNV RILASGGTAR MIRDAGFPIE DVSAITHAPE 60
61 MLGGRVKTLH PAVHGGILAR DIDSDEKDLK EQHIEKVDYV VCNLYPFKET VAKVGVTIPE 120
121 AVEEIDIGGV TLLRAAAKNH ARVTILSDPK DYSEFLSELS SNGEISQDLR NRLALKAFEH 180
181 TADYDAAISD FFRKQYSEGQ AQITLRYGAN PHQKPAQAYV SQQDSLPFKV LCGSPGYINL 240
241 LDALNSWPLV KELSASLNLP AAASFKHVSP AGAAVGIPLS DVEKQVYFVA DIENLSPLAC 300
301 AYARARGADR MSSFGDWIAL SNIVDVPTAK IISREVSDGV IAPGYEPEAL AILSKKKGGK 360
361 YCILQIDPNY VPEAVERRQV YGVTLEQKRN DAIINQSTFK EIVSQNKNLT EQAIIDLTVA 420
421 TIAIKYTQSN SVCYARNGMV VGLGAGQQSR IHCTRLAGDK ADNWWFRQHP RVLEIKWAKG 480
481 VKRPEKSNAI DLFVTGQIPT EEPELSEYQS KFEEIPKPFT PEERKEWLSK LTNVSLSSDA 540
541 FFPFPDNVYR AVKSGVKYIA APSGSVMDKV VFSAADSFDL VYVENPIRLF HH |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
ADE16 ADE17 |
| View Details | Krogan NJ, et al. (2006) |
|
ADE16 ADE17 SLX1 |
| View Details | Ho Y, et al. (2002) |
|
ADE17 MDH1 PGM2 PRP6 SEC53 SSZ1 STE4 TPS1 |
| View Details | Qiu et al. (2008) |
|
ADE16 ADE17 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
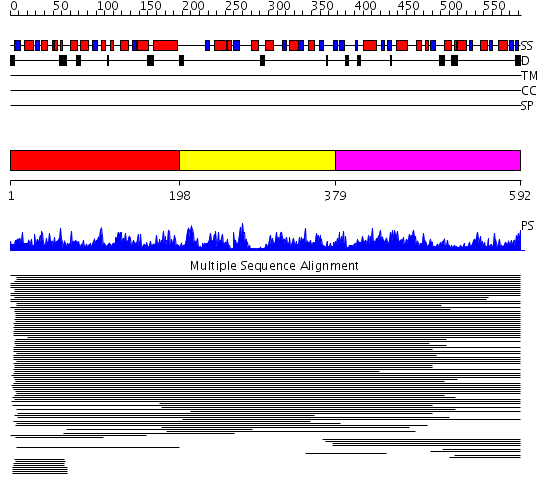
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..197] | 11000.0 | IMP cyclohydrolase domain of bifunctional purine biosynthesis enzyme ATIC; AICAR transformylase domain of bifunctional purine biosynthesis enzyme ATIC | |
| 2 | View Details | [198..378] | 11000.0 | IMP cyclohydrolase domain of bifunctional purine biosynthesis enzyme ATIC; AICAR transformylase domain of bifunctional purine biosynthesis enzyme ATIC | |
| 3 | View Details | [379..592] | 11000.0 | IMP cyclohydrolase domain of bifunctional purine biosynthesis enzyme ATIC; AICAR transformylase domain of bifunctional purine biosynthesis enzyme ATIC |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)