













| General Information: |
|
| Name(s) found: |
PMS1 /
YNL082W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 873 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear chromosome
[TAS |
| Biological Process: |
meiotic mismatch repair
[IMP mismatch repair [TAS meiosis [TAS |
| Molecular Function: |
ATPase activity
[IDA DNA binding [IDA ATP binding [TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTQIHQINDI DVHRITSGQV ITDLTTAVKE LVDNSIDANA NQIEIIFKDY GLESIECSDN 60
61 GDGIDPSNYE FLALKHYTSK IAKFQDVAKV QTLGFRGEAL SSLCGIAKLS VITTTSPPKA 120
121 DKLEYDMVGH ITSKTTTSRN KGTTVLVSQL FHNLPVRQKE FSKTFKRQFT KCLTVIQGYA 180
181 IINAAIKFSV WNITPKGKKN LILSTMRNSS MRKNISSVFG AGGMRGLEEV DLVLDLNPFK 240
241 NRMLGKYTDD PDFLDLDYKI RVKGYISQNS FGCGRNSKDR QFIYVNKRPV EYSTLLKCCN 300
301 EVYKTFNNVQ FPAVFLNLEL PMSLIDVNVT PDKRVILLHN ERAVIDIFKT TLSDYYNRQE 360
361 LALPKRMCSQ SEQQAQKRLK TEVFDDRSTT HESDNENYHT ARSESNQSNH AHFNSTTGVI 420
421 DKSNGTELTS VMDGNYTNVT DVIGSECEVS VDSSVVLDEG NSSTPTKKLP SIKTDSQNLS 480
481 DLNLNNFSNP EFQNITSPDK ARSLEKVVEE PVYFDIDGEK FQEKAVLSQA DGLVFVDNEC 540
541 HEHTNDCCHQ ERRGSTDTEQ DDEADSIYAE IEPVEINVRT PLKNSRKSIS KDNYRSLSDG 600
601 LTHRKFEDEI LEYNLSTKNF KEISKNGKQM SSIISKRKSE AQENIIKNKD ELEDFEQGEK 660
661 YLTLTVSKND FKKMEVVGQF NLGFIIVTRK VDNKYDLFIV DQHASDEKYN FETLQAVTVF 720
721 KSQKLIIPQP VELSVIDELV VLDNLPVFEK NGFKLKIDEE EEFGSRVKLL SLPTSKQTLF 780
781 DLGDFNELIH LIKEDGGLRR DNIRCSKIRS MFAMRACRSS IMIGKPLNKK TMTRVVHNLS 840
841 ELDKPWNCPH GRPTMRHLME LRDWSSFSKD YEI |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
MLH1 PMS1 |
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
MSH2 MSH3 PMS1 |
| View Details | Krogan NJ, et al. (2006) |
|
NSA1 PMS1 RDR1 |
| View Details | Gavin AC, et al. (2006) |

|
MLH1 PMS1 |
| View Details | Gavin AC, et al. (2002) |

|
CMR1 DNA2 HDA1 HDA2 HDA3 HSC82 KAP95 LCD1 MEC1 MGM101 MLH1 MPH1 MSH2 MSH3 MSH6 OLA1 PGK1 PMS1 PRO2 RAD16 RAD52 RFA1 RFA2 RFA3 RIM1 RNH1 RSC8 RVB2 SGS1 SHS1 TIF1, TIF2 TOP3 VAS1 VPS1 YHR122W |
| View Details | Qiu et al. (2008) |

|
MSH2 PMS1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
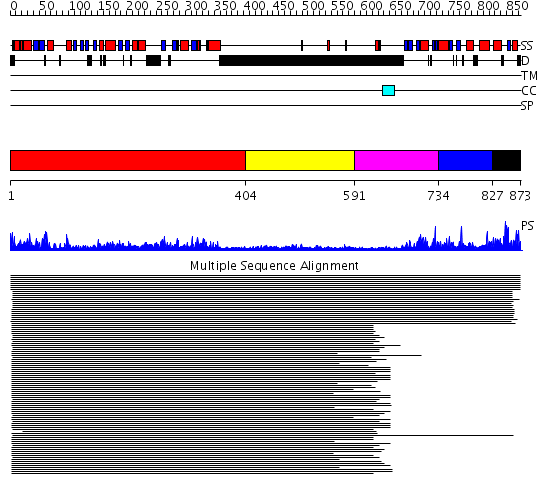
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..403] | 68.39794 | DNA mismatch repair protein PMS2 | |
| 2 | View Details | [404..590] | 4.434992 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [591..733] | 1.139986 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [734..826] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [827..873] | 3.062993 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)