













| General Information: |
|
| Name(s) found: |
RDR1 /
YOR380W
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 546 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[TAS |
| Biological Process: |
response to xenobiotic stimulus
[IMP |
| Molecular Function: |
transcription factor activity
[IGI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASPGSTALP HKRQRVRKAC VPCRERKRKC NGKSPCEMCV AYGYVCHYID GRVPSASPQV 60
61 QQVGETSPDT ESRPFVLPGI HRNEQPQPIN TQNVTSQNIV DPTKSRYTIQ HSAVAFPRCL 120
121 GLELRSTNPP RLHSFAWHCG IRPEENPNSH VLLSDLVTKE EYYRISKVYF SVVHPIFDVV 180
181 NPEQLAKNVE KYWDGDVKTL EYGAVIAGVI ALGSFFMGSL GHPREMDIVQ YAKGILDDPT 240
241 FSRIPTVEQV SAWVLRTIYL RATSRPHVAW LASCVTIHLS EAIGLHHEID REDIAISNNV 300
301 PPKRTTVVSE HTRRLFWCAW SINTILSYDY GRSSVTLNRI TCKPVKETDG NFTAHLVALA 360
361 HLIPQDSVNA NAAQLLQALA AVHESPNAHP FLSLTKGDIC LSLYRRLRLL NHILDKNVVL 420
421 QIIDIGNTAL SAAYALVKLD QAWWNVLSTS FQYVCVLLAI DTPESLSHVA TAMKTLDNIT 480
481 QILGTRIAFE AQKTAKLLLE DSMKKKRQEI QQLEQATHQR SNLETTHLLD IDWDALLDPS 540
541 DTLNFM |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |

|
NSA1 PMS1 RDR1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
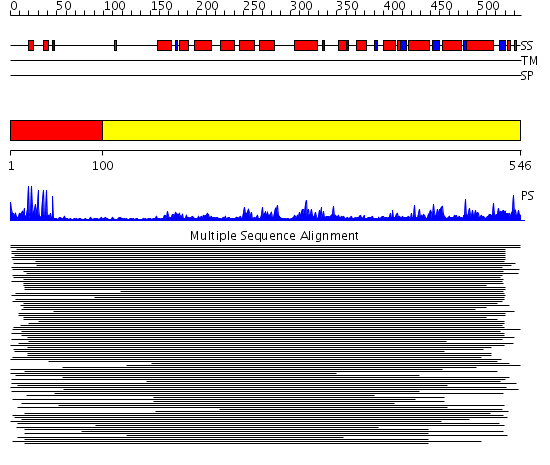
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..99] | 36.0 | PUT3 | |
| 2 | View Details | [100..546] | 9.232982 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [223..351] | 19.568636 | No description for PF04082.9 was found. No confident structure predictions are available. | |
| 4 | View Details | [352..546] | 4.393996 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.74 |
Source: Reynolds et al. (2008)