













| General Information: |
|
| Name(s) found: |
PDX1 /
YGR193C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 410 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrial pyruvate dehydrogenase complex
[TAS mitochondrion [IDA |
| Biological Process: |
acetyl-CoA biosynthetic process from pyruvate
[TAS |
| Molecular Function: |
protein binding
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLSAISKVST LKSCTRYLTK CNYHASAKLL AVKTFSMPAM SPTMEKGGIV SWKYKVGEPF 60
61 SAGDVILEVE TDKSQIDVEA LDDGKLAKIL KDEGSKDVDV GEPIAYIADV DDDLATIKLP 120
121 QEANTANAKS IEIKKPSADS TEATQQHLKK ATVTPIKTVD GSQANLEQTL LPSVSLLLAE 180
181 NNISKQKALK EIAPSGSNGR LLKGDVLAYL GKIPQDSVNK VTEFIKKNER LDLSNIKPIQ 240
241 LKPKIAEQAQ TKAADKPKIT PVEFEEQLVF HAPASIPFDK LSESLNSFMK EAYQFSHGTP 300
301 LMDTNSKYFD PIFEDLVTLS PREPRFKFSY DLMQIPKANN MQDTYGQEDI FDLLTGSDAT 360
361 ASSVRPVEKN LPEKNEYILA LNVSVNNKKF NDAEAKAKRF LDYVRELESF |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
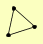
|
LAT1 PDA1 PDX1 |
| View Details | (MIPS) Mewes HW, et al. (2004) |
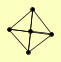
|
LAT1 LPD1 PDA1 PDB1 PDX1 |
| View Details | Gavin AC, et al. (2006) |
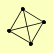
|
LAT1 PDA1 PDB1 PDX1 |
| View Details | Gavin AC, et al. (2002) |
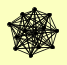
|
APM1 CYM1 DOA4 DUR1,2 HSP42 LAT1 LPD1 PDA1 PDB1 PDX1 PRP46 RPN11 TCM62 |
| View Details | Ho Y, et al. (2002) |

|
ATP3 AVO1 CCT5 CLU1 DHH1 DPB2 FUN12 GAL7 GPH1 HHF1, HHF2 IDH1 ILV2 MET16 MKK2 NOG2 NPA3 PDX1 PHO85 PUF3 RAD16 RPN12 SIS1 SLX1 SRP54 STI1 TEM1 TFC7 VMA8 YER077C YGR266W YKR051W YKU80 YLR271W YPR003C |
| View Details | Qiu et al. (2008) |
|
LAT1 LPD1 PDA1 PDB1 PDX1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | Sample boi 1 with ha tag from october 2005 | McCusker D, et al (2007) | |
| View Run | sample: hx3 | Hao Xu, et al. (2010) | |
| View Run | Sample: HX11 - both fusion and trans-SNARE complex formation take place. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | Sample: HX13 - trans-SNARE complex forms but fusion is blocked. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX14 - mixture in detergent (control). | Hao Xu, et al. (2010) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
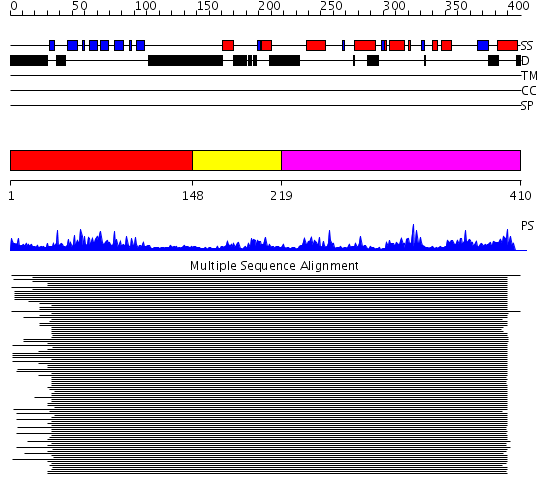
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..147] | 163.522879 | Ipoyl domain of dihydrolipoamide acetyltransferase | |
| 2 | View Details | [148..218] | 4.045757 | E3-binding domain of dihydrolipoamide succinyltransferase | |
| 3 | View Details | [219..410] | 39.39794 | Dihydrolipoamide succinyltransferase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)