













| General Information: |
|
| Name(s) found: |
BUB1 /
YGR188C
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1021 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA condensed nuclear chromosome, centromeric region [IDA condensed nuclear chromosome kinetochore [IDA kinetochore [IDA |
| Biological Process: |
mitotic cell cycle spindle assembly checkpoint
[TAS protein amino acid phosphorylation [IDA |
| Molecular Function: |
protein binding
[IPI protein kinase activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNLDLGSTVR GYESDKDTFP QSKGVSSSQK EQHSQLNQTK IAYEQRLLND LEDMDDPLDL 60
61 FLDYMIWIST SYIEVDSESG QEVLRSTMER CLIYIQDMET YRNDPRFLKI WIWYINLFLS 120
121 NNFHESENTF KYMFNKGIGT KLSLFYEEFS KLLENAQFFL EAKVLLELGA ENNCRPYNRL 180
181 LRSLSNYEDR LREMNIVENQ NSVPDSRERL KGRLIYRTAP FFIRKFLTSS LMTDDKENRA 240
241 NLNSNVGVGK SAPNVYQDSI VVADFKSETE RLNLNSSKQP SNQRLKNGNK KTSIYADQKQ 300
301 SNNPVYKLIN TPGRKPERIV FNFNLIYPEN DEEFNTEEIL AMIKGLYKVQ RRGKKHTEDY 360
361 TSDKNRKKRK LDVLVERRQD LPSSQPPVVP KSTRIEVFKD DDNPSQSTHH KNTQVQVQTT 420
421 TSILPLKPVV DGNLAHETPV KPSLTSNASR SPTVTAFSKD AINEVFSMFN QHYSTPGALL 480
481 DGDDTTTSKF NVFENFTQEF TAKNIEDLTE VKDPKQETVS QQTTSTNETN DRYERLSNSS 540
541 TRPEKADYMT PIKETTETDV VPIIQTPKEQ IRTEDKKSGD NTETQTQLTS TTIQSSPFLT 600
601 QPEPQAEKLL QTAEHSEKSK EHYPTIIPPF TKIKNQPPVI IENPLSNNLR AKFLSEISPP 660
661 LFQYNTFYNY NQELKMSSLL KKIHRVSRNE NKNPIVDFKK TGDLYCIRGE LGEGGYATVY 720
721 LAESSQGHLR ALKVEKPASV WEYYIMSQVE FRLRKSTILK SIINASALHL FLDESYLVLN 780
781 YASQGTVLDL INLQREKAID GNGIMDEYLC MFITVELMKV LEKIHEVGII HGDLKPDNCM 840
841 IRLEKPGEPL GAHYMRNGED GWENKGIYLI DFGRSFDMTL LPPGTKFKSN WKADQQDCWE 900
901 MRAGKPWSYE ADYYGLAGVI HSMLFGKFIE TIQLQNGRCK LKNPFKRYWK KEIWGVIFDL 960
961 LLNSGQASNQ ALPMTEKIVE IRNLIESHLE QHAENHLRNV ILSIEEELSH FQYKGKPSRR1020
1021 F |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample boi 1 with ha tag from october 2005 | McCusker D, et al (2007) | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
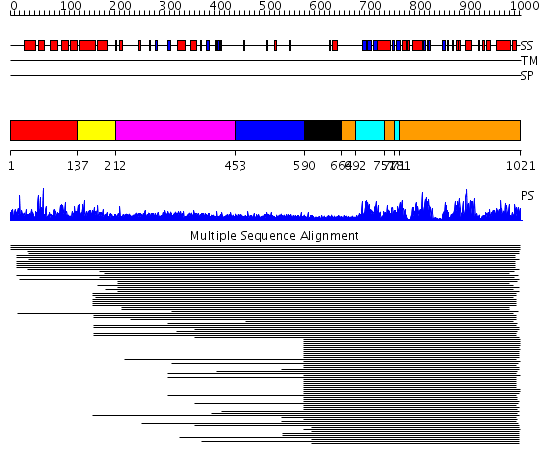
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..136] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [137..211] | 3.69897 | Effector domain of the protein kinase pkn/prk1 | |
| 3 | View Details | [212..452] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [453..589] | 11.0 | Photoreceptor phy3 flavin-binding domain, lov2 | |
| 5 | View Details | [590..663] | 6.221849 | C2 domain from protein kinase c (beta) | |
| 6 | View Details | [664..691] [751..770] [781..1021] |
105.0 | cAMP-dependent PK, catalytic subunit | |
| 7 | View Details | [692..750] [771..780] |
105.0 | cAMP-dependent PK, catalytic subunit |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)