













| General Information: |
|
| Name(s) found: |
SPT2 /
YER161C
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 333 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
negative regulation of transcription from RNA polymerase II promoter
[IMP RNA polyadenylation [IGI chromatin organization [IMP RNA elongation from RNA polymerase II promoter [IGI |
| Molecular Function: |
DNA binding
[IDA DNA secondary structure binding [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSFLSKLSQI RKSTTASKAQ VQDPLPKKND EEYSLLPKNY IRDEDPAVKR LKELRRQELL 60
61 KNGALAKKSG VKRKRGTSSG SEKKKIERND DDEGGLGIRF KRSIGASHAP LKPVVRKKPE 120
121 PIKKMSFEEL MKQAENNEKQ PPKVKSSEPV TKERPHFNKP GFKSSKRPQK KASPGATLRG 180
181 VSSGGNSIKS SDSPKPVKLN LPTNGFAQPN RRLKEKLESR KQKSRYQDDY DEEDNDMDDF 240
241 IEDDEDEGYH SKSKHSNGPG YDRDEIWAMF NRGKKRSEYD YDELEDDDME ANEMEILEEE 300
301 EMARKMARLE DKREEAWLKK HEEEKRRRKK GIR |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
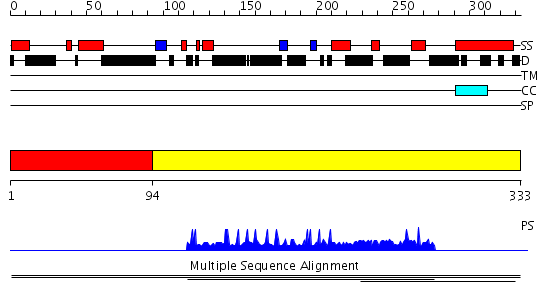
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..93] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [94..333] | 1.002939 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)