













| General Information: |
|
| Name(s) found: |
SPC110 /
YDR356W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 944 amino acids |
Gene Ontology: |
|
| Cellular Component: |
inner plaque of spindle pole body
[IDA central plaque of spindle pole body [IDA |
| Biological Process: |
microtubule nucleation
[IPI |
| Molecular Function: |
structural constituent of cytoskeleton
[IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDEASHLPNG SLKNMEFTPV GFIKSKRNTT QTQVVSPTKV PNANNGDENE GPVKKRQRRS 60
61 IDDTIDSTRL FSEASQFDDS FPEIKANIPP SPRSGNVDKS RKRNLIDDLK KDVPMSQPLK 120
121 EQEVREHQMK KERFDRALES KLLGKRHITY ANSDISNKEL YINEIKSLKH EIKELRKEKN 180
181 DTLNNYDTLE EETDDLKNRL QALEKELDAK NKIVNSRKVD DHSGCIEERE QMERKLAELE 240
241 RKLKTVKDQV LELENNSDVQ SLKLRSKEDE LKNLMNELNE LKSNAEEKDT QLEFKKNELR 300
301 KRTNELNELK IKSDEMDLQL KQKQNESKRL KDELNELETK FSENGSQSSA KENELKMLKN 360
361 KIAELEEEIS TKNSQLIAKE GKLASLMAQL TQLESKLNQR DSQLGSREEE LKKTNDKLQK 420
421 DIRIAREETV SKDERIIDLQ KKVKQLENDL FVIKKTHSES KTITDNELES KDKLIKILEN 480
481 DLKVAQEKYS KMEKELKERE FNYKISESKL EDEKTTLNEK ISNLAAENSQ LKNKIEDNST 540
541 ATHHMKENYE KQLESLRKDI EEYKESAKDS EDKIEELKIR IAENSAKVSE KRSKDIKQKD 600
601 EQISDLTQNL KLQEDEISSL KSIIDRYKKD FNQLKSEQSN IQHDLNLQIL NLENKLIESE 660
661 DELKSLRDSQ KIEIENWKRK YNNLSLENDR LLTEKESASD KEREISILNR KLDEMDKEKW 720
721 NLQESKEKYK RELQKVITAN DRLRREKEEL NENSNNIRIM EDKMTRIKKN YLSEITSLQE 780
781 ENRRLEERLI LNERRKDNDS TMQLNDIISY YKLKYHSEVR HNNDLKVIND YLNKVLALGT 840
841 RRLRLDTRKG EHSLNISLPD DDELDRDYYN SHVYTRYHDY EYPLRFNLNR RGPYFERRLS 900
901 FKTVALLVLA CVRMKRIAFY RRSDDNRLRI LRDRIESSSG RISW |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | (MIPS) Mewes HW, et al. (2004) |
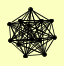
|
BBP1 CDC31 CMD1 CNM67 KAR1 NUD1 SPC105 SPC110 SPC24 SPC25 SPC29 SPC42 SPC72 |
| View Details | Gavin AC, et al. (2002) |
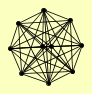
|
CDC48 GIM3 GIM5 PAC10 PFK1 SPC110 SPC97 SPC98 TUB4 YKE2 |
| View Details | Ho Y, et al. (2002) |
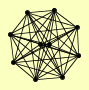
|
CMD1 CMP2 HUL5 MLC1 MYO2 MYO3 MYO4 MYO5 SHE4 SPC110 |
| View Details | Qiu et al. (2008) |
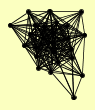
|
BBP1 BIK1 CDC31 CMD1 CNM67 KAR1 MPS2 NDC1 NUD1 NUF2 SLK19 SPC105 SPC110 SPC19 SPC24 SPC25 SPC29 SPC42 SPC72 SPC97 SPC98 STU1 STU2 TID3 TUB4 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #33 Asynchronous SPB prep | Keck JM, et al. (2011) | |
| View Run | #19 Asynchronous Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) | |
| View Run | #21 Asynchronous Prep2-IMAC Phosphopeptide enrichment A1 | Keck JM, et al. (2011) | |
| View Run | #22 Asynchronous Prep2-IMAC Phosphopeptide enrichment B1-2 | Keck JM, et al. (2011) | |
| View Run | #34 Asynchronous SPB prep | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #35 Asynchronous SPB prep | Keck JM, et al. (2011) | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) | |
| View Run | #04 Alpha Factor Prep2 | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #14 Mitotic Prep2-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #13 Mitotic Prep2-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #02 Alpha Factor Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #01 Alpha factor Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #28 Asynchronous Prep5-TiO2 Phosphopeptide Enrichment | Keck JM, et al. (2011) | |
| View Run | #06 Alpha Factor Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #08 Alpha Factor Prep4-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #16 Mitotic-New search criteria | Keck JM, et al. (2011) | |
| View Run | #15 Mitotic 2nd-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #19b Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) | |
| View Run | #05 Alpha Factor Prep2-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #07 Alpha Factor Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #09 Alpha Factor Prep4-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #17 Mitotic Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #18 Mitotic Prep4-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #19a Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Sundin BA, et al. (2004) |
| View Data |
|
Sundin BA, et al. (2004) |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
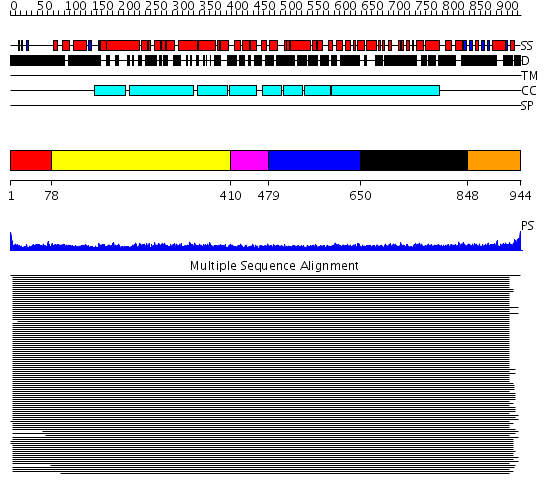
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..77] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [78..409] | 63.68867 | Heavy meromyosin subfragment | |
| 3 | View Details | [410..478] | 24.39794 | Tropomyosin | |
| 4 | View Details | [479..649] | 14.1 | alpha-actinin | |
| 5 | View Details | [650..847] | 14.1 | alpha-actinin | |
| 6 | View Details | [848..944] | N/A | Confident ab initio structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.86 |
Source: Reynolds et al. (2008)