













| General Information: |
|
| Name(s) found: |
CDC48 /
YDL126C
[SGD]
|
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 835 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endoplasmic reticulum membrane
[IDA nucleus [IDA microsome [IDA cytosol [IDA |
| Biological Process: |
vesicle fusion
[IDA apoptosis [TAS protein transport [IDA ER-associated protein catabolic process [IMP cell cycle [IMP ubiquitin-dependent protein catabolic process [IDA |
| Molecular Function: |
ATPase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGEEHKPLLD ASGVDPREED KTATAILRRK KKDNMLLVDD AINDDNSVIA INSNTMDKLE 60
61 LFRGDTVLVK GKKRKDTVLI VLIDDELEDG ACRINRVVRN NLRIRLGDLV TIHPCPDIKY 120
121 ATRISVLPIA DTIEGITGNL FDVFLKPYFV EAYRPVRKGD HFVVRGGMRQ VEFKVVDVEP 180
181 EEYAVVAQDT IIHWEGEPIN REDEENNMNE VGYDDIGGCR KQMAQIREMV ELPLRHPQLF 240
241 KAIGIKPPRG VLMYGPPGTG KTLMARAVAN ETGAFFFLIN GPEVMSKMAG ESESNLRKAF 300
301 EEAEKNAPAI IFIDEIDSIA PKRDKTNGEV ERRVVSQLLT LMDGMKARSN VVVIAATNRP 360
361 NSIDPALRRF GRFDREVDIG IPDATGRLEV LRIHTKNMKL ADDVDLEALA AETHGYVGAD 420
421 IASLCSEAAM QQIREKMDLI DLDEDEIDAE VLDSLGVTMD NFRFALGNSN PSALRETVVE 480
481 SVNVTWDDVG GLDEIKEELK ETVEYPVLHP DQYTKFGLSP SKGVLFYGPP GTGKTLLAKA 540
541 VATEVSANFI SVKGPELLSM WYGESESNIR DIFDKARAAA PTVVFLDELD SIAKARGGSL 600
601 GDAGGASDRV VNQLLTEMDG MNAKKNVFVI GATNRPDQID PAILRPGRLD QLIYVPLPDE 660
661 NARLSILNAQ LRKTPLEPGL ELTAIAKATQ GFSGADLLYI VQRAAKYAIK DSIEAHRQHE 720
721 AEKEVKVEGE DVEMTDEGAK AEQEPEVDPV PYITKEHFAE AMKTAKRSVS DAELRRYEAY 780
781 SQQMKASRGQ FSNFNFNDAP LGTTATDNAN SNNSAPSGAG AAFGSNAEED DDLYS |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
CDC48 DOA1 NPL4 SHP1 UFD1 UFD2 YDR049W |
| View Details | Krogan NJ, et al. (2006) |

|
BFA1 CDC48 DOA1 FBP1 MRM1 NDT80 NPL4 OTU1 RDS2 SHP1 TEM1 TUP1 UBX4 UBX5 UBX7 UFD1 UFD2 YDR049W YGL108C YJR098C YNL155W YOR342C |
| View Details | Gavin AC, et al. (2006) |
|
CDC48 NPL4 SHP1 UFD1 YDR049W |
| View Details | Gavin AC, et al. (2002) |
|
BNI1 CDC48 DOA1 GIM3 GIM5 KRS1 MRPS28 NPL4 PAC10 PDC1 PFK1 RAI1 REG1 RPA135 RPA190 RPO31 SHP1 SPC110 SPC97 SPC98 TFB4 TUB4 UFD1 YDR049W YKE2 |
The following runs contain data for this protein:
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
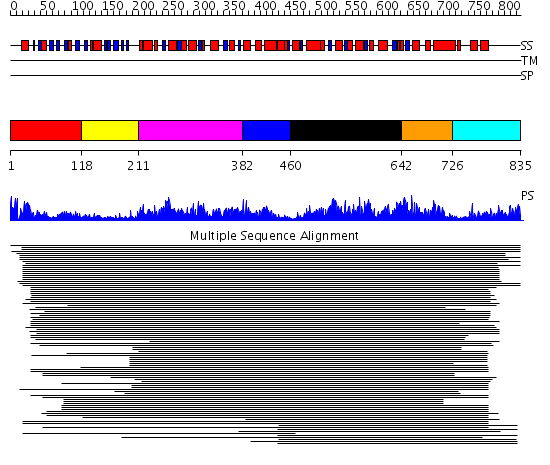
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..117] | 10138.0 | Membrane fusion atpase p97 N-terminal domain , P97-Nn; Membrane fusion atpase p97, D1 domain; Membrane fusion atpase p97 domain 2, P97-Nc | |
| 2 | View Details | [118..210] | 10138.0 | Membrane fusion atpase p97 N-terminal domain , P97-Nn; Membrane fusion atpase p97, D1 domain; Membrane fusion atpase p97 domain 2, P97-Nc | |
| 3 | View Details | [211..381] | 10138.0 | Membrane fusion atpase p97 N-terminal domain , P97-Nn; Membrane fusion atpase p97, D1 domain; Membrane fusion atpase p97 domain 2, P97-Nc | |
| 4 | View Details | [382..459] | 10138.0 | Membrane fusion atpase p97 N-terminal domain , P97-Nn; Membrane fusion atpase p97, D1 domain; Membrane fusion atpase p97 domain 2, P97-Nc | |
| 5 | View Details | [460..641] | 64.751666 | Hexamerization domain of N-ethylmalemide-sensitive fusion (NSF) protein | |
| 6 | View Details | [642..725] | 11.93 | Hexamerization domain of N-ethylmalemide-sensitive fusion (NSF) protein | |
| 7 | View Details | [726..835] | 4.430994 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)