













| General Information: |
|
| Name(s) found: |
NDT80 /
YHR124W
[SGD]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 627 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear chromosome
[IDA |
| Biological Process: |
transcription
[IGI meiosis [IEP |
| Molecular Function: |
transcription factor activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNEMENTDPV LQDDLVSKYE RELSTEQEED TPVILTQLNE DGTTSNYFDK RKLKIAPRST 60
61 LQFKVGPPFE LVRDYCPVVE SHTGRTLDLR IIPRIDRGFD HIDEEWVGYK RNYFTLVSTF 120
121 ETANCDLDTF LKSSFDLLVE DSSVESRLRV QYFAIKIKAK NDDDDTEINL VQHTAKRDKG 180
181 PQFCPSVCPL VPSPLPKHQI IREASNVRNI TKMKKYDSTF YLHRDHVNYE EYGVDSLLFS 240
241 YPEDSIQKVA RYERVQFASS ISVKKPSQQN KHFSLHVILG AVVDPDTFHG ENPGIPYDEL 300
301 ALKNGSKGMF VYLQEMKTPP LIIRGRSPSN YASSQRITVR TPSSVNSSQN STKRKMPSMA 360
361 QPLNESCLNA RPSKRRSKVA LGAPNSGASI SPIKSRQSTP MEASKENEDP FFRPNKRVET 420
421 LEHIQNKLGA LKNQCPDSSL KYPSSSSRGM EGCLEKEDLV YSSSFSVNMK QIELKPARSF 480
481 EHENIFKVGS LAFKKINELP HENYDITIEK KSMEQNYLRP EIGSRSECKT SYGNELSLSN 540
541 ISFSILPNSA ENFHLETALF PATEEDVPRT FSRILETGSF QNYYQKMDAE NADRVYSKGV 600
601 KLIASGTLPS GIFNREELFE EDSFYKY |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
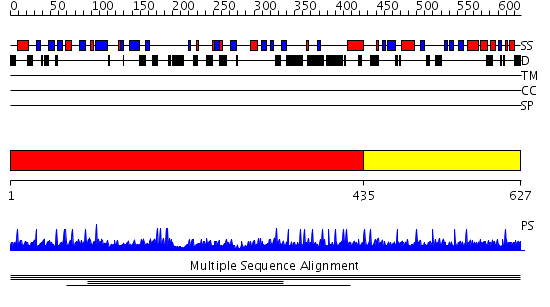
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..434] | 1.003997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [435..627] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [524..627] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)