













| General Information: |
|
| Name(s) found: |
SIF2 /
YBR103W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 535 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IPI histone deacetylase complex [IPI |
| Biological Process: |
negative regulation of transcription from RNA polymerase II promoter
[IPI histone deacetylation [IDA chromatin silencing at telomere [IGI negative regulation of meiosis [IPI |
| Molecular Function: |
transcription corepressor activity
[IDA NAD-dependent histone deacetylase activity [IPI NAD-independent histone deacetylase activity [IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSITSEELNY LIWRYCQEMG HEVSALALQD ETRVLEFDEK YKEHIPLGTL VNLVQRGILY 60
61 TESELMVDSK GDISALNEHH LSEDFNLVQA LQIDKEKFPE ISSEGRFTLE TNSESNKAGE 120
121 DGASTVERET QEDDTNSIDS SDDLDGFVKI LKEIVKLDNI VSSTWNPLDE SILAYGEKNS 180
181 VARLARIVET DQEGKKYWKL TIIAELRHPF ALSASSGKTT NQVTCLAWSH DGNSIVTGVE 240
241 NGELRLWNKT GALLNVLNFH RAPIVSVKWN KDGTHIISMD VENVTILWNV ISGTVMQHFE 300
301 LKETGGSSIN AENHSGDGSL GVDVEWVDDD KFVIPGPKGA IFVYQITEKT PTGKLIGHHG 360
361 PISVLEFNDT NKLLLSASDD GTLRIWHGGN GNSQNCFYGH SQSIVSASWV GDDKVISCSM 420
421 DGSVRLWSLK QNTLLALSIV DGVPIFAGRI SQDGQKYAVA FMDGQVNVYD LKKLNSKSRS 480
481 LYGNRDGILN PLPIPLYASY QSSQDNDYIF DLSWNCAGNK ISVAYSLQEG SVVAI |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
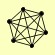
|
CPR1 HOS2 HOS4 SET3 SIF2 SNT1 |
| View Details | Krogan NJ, et al. (2006) |
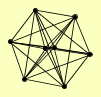
|
AKL1 HOS4 HST1 RFM1 SET3 SIF2 SNT1 SUM1 |
| View Details | Gavin AC, et al. (2006) |
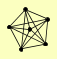
|
CPR1 HOS2 HOS4 SET3 SIF2 SNT1 |
| View Details | Gavin AC, et al. (2002) |

|
BLM10 CDC55 CIN1 CPR1 ERG13 GUS1 HHF1, HHF2 HOS2 HOS4 HST1 IML1 KAP95 KEL1 LTE1 MYO5 PFK1 PPH21 PPH22 PRE1 PRE10 PRE2 PRE4 PRE5 PRE6 PRE7 PRE8 PRE9 PUP1 PUP2 PUP3 RRD2 RTS1 RTS3 RVB2 SCL1 SET3 SIF2 SNF2 SNT1 SRP1 TDH2 TDH3 TEF4 TPD3 YBL104C YOR1 YRA1 ZDS1 ZDS2 |
| View Details | Ho Y, et al. (2002) |
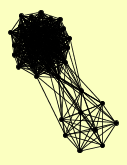
|
AVT2 DIS3 EAF3 FIP1 HAS1 HOS4 HPR1 KAP95 MES1 MFT1 NAM8 NOP6 NUP1 NUP2 NUP60 PAP1 PCT1 REB1 RNT1 RRP4 RRP43 RRP6 RTT103 SIF2 SIN3 SNT1 SNU56 SRP1 STO1 TFP1 THO2 TRA1 TRM3 UME1 UTP21 YMR155W YPR090W YRF1-3, YRF1-7 ZDS2 |
| View Details | Qiu et al. (2008) |
|
HDA3 HOS2 HOS4 HST1 RPD3 RXT2 SDS3 SET3 SIF2 SNT1 STB2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Uetz P, et al. (2000) |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
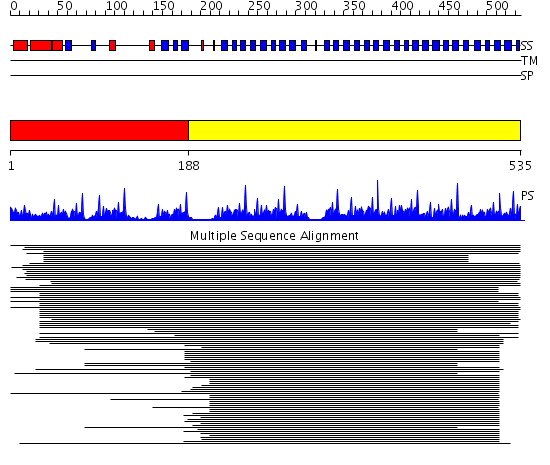
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..187] | 4.514997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [188..535] | 130.29073 | beta1-subunit of the signal-transducing G protein heterotrimer |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)