













| General Information: |
|
| Name(s) found: |
SHP1 /
YBL058W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 423 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
ascospore formation
[IMP proteasomal ubiquitin-dependent protein catabolic process [IMP glycogen metabolic process [IMP telomere maintenance [IMP |
| Molecular Function: |
protein phosphatase type 1 regulator activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAEIPDETIQ QFMALTNVSH NIAVQYLSEF GDLNEALNSY YASQTDDQKD RREEAHWNRQ 60
61 QEKALKQEAF STNSSNKAIN TEHVGGLCPK PGSSQGSNEY LKRKGSTSPE PTKGSSRSGS 120
121 GNNSRFMSFS DMVRGQADDD DEDQPRNTFA GGETSGLEVT DPSDPNSLLK DLLEKARRGG 180
181 QMGAENGFRD DEDHEMGANR FTGRGFRLGS TIDAADEVVE DNTSQSQRRP EKVTREITFW 240
241 KEGFQVADGP LYRYDDPANS FYLSELNQGR APLKLLDVQF GQEVEVNVYK KLDESYKAPT 300
301 RKLGGFSGQG QRLGSPIPGE SSPAEVPKNE TPAAQEQPMP DNEPKQGDTS IQIRYANGKR 360
361 EVLHCNSTDT VKFLYEHVTS NANTDPSRNF TLNYAFPIKP ISNDETTLKD ADLLNSVVVQ 420
421 RWA |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
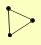
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
CDC48 NPL4 SHP1 |
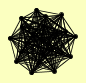
| View Details | Krogan NJ, et al. (2006) |
|
BFA1 CDC48 DOA1 FBP1 MRM1 NDT80 NPL4 OTU1 RDS2 SHP1 TEM1 TUP1 UBX4 UBX5 UBX7 UFD1 UFD2 YDR049W YGL108C YJR098C YNL155W YOR342C |
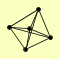
| View Details | Gavin AC, et al. (2006) |
|
CDC48 NPL4 SHP1 UFD1 YDR049W |
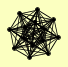
| View Details | Gavin AC, et al. (2002) |
|
BNI1 CDC48 DOA1 MRPS28 NPL4 PDC1 RAI1 REG1 RPA135 RPA190 RPO31 SHP1 UFD1 YDR049W |
| View Details | Ho Y, et al. (2002) |
|
DSK2 GND1 HHF1, HHF2 HMF1 HTB2 HTZ1 NPL4 RAD16 RAD23 RAD7 RNQ1 SHP1 TDP1 TMA29 TSA2 TSL1 UBI4 UFD2 YDR049W |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
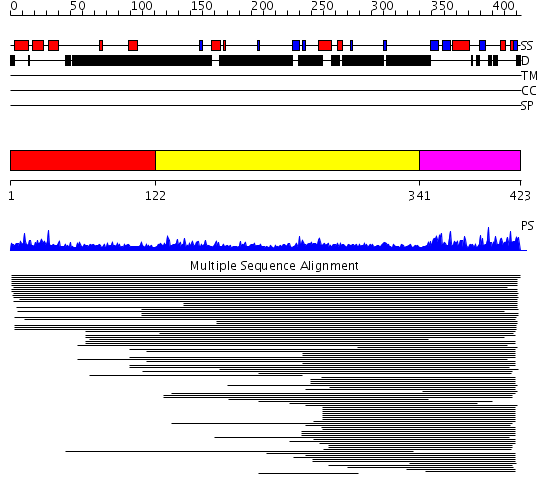
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..121] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [122..340] | 1.067995 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [341..423] | 77.071898 | p47 | |
| 4 | View Details | [313..423] | 34.221849 | Crystal structure of AAA ATPase p97/VCP ND1 in complex with p47 C |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)