













| Protein: | TMA19 |
| From Publication: | Ho Y. et al. (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002 Jan 10;415(6868):180-3. |
| Complexes containing TMA19: | 7 |
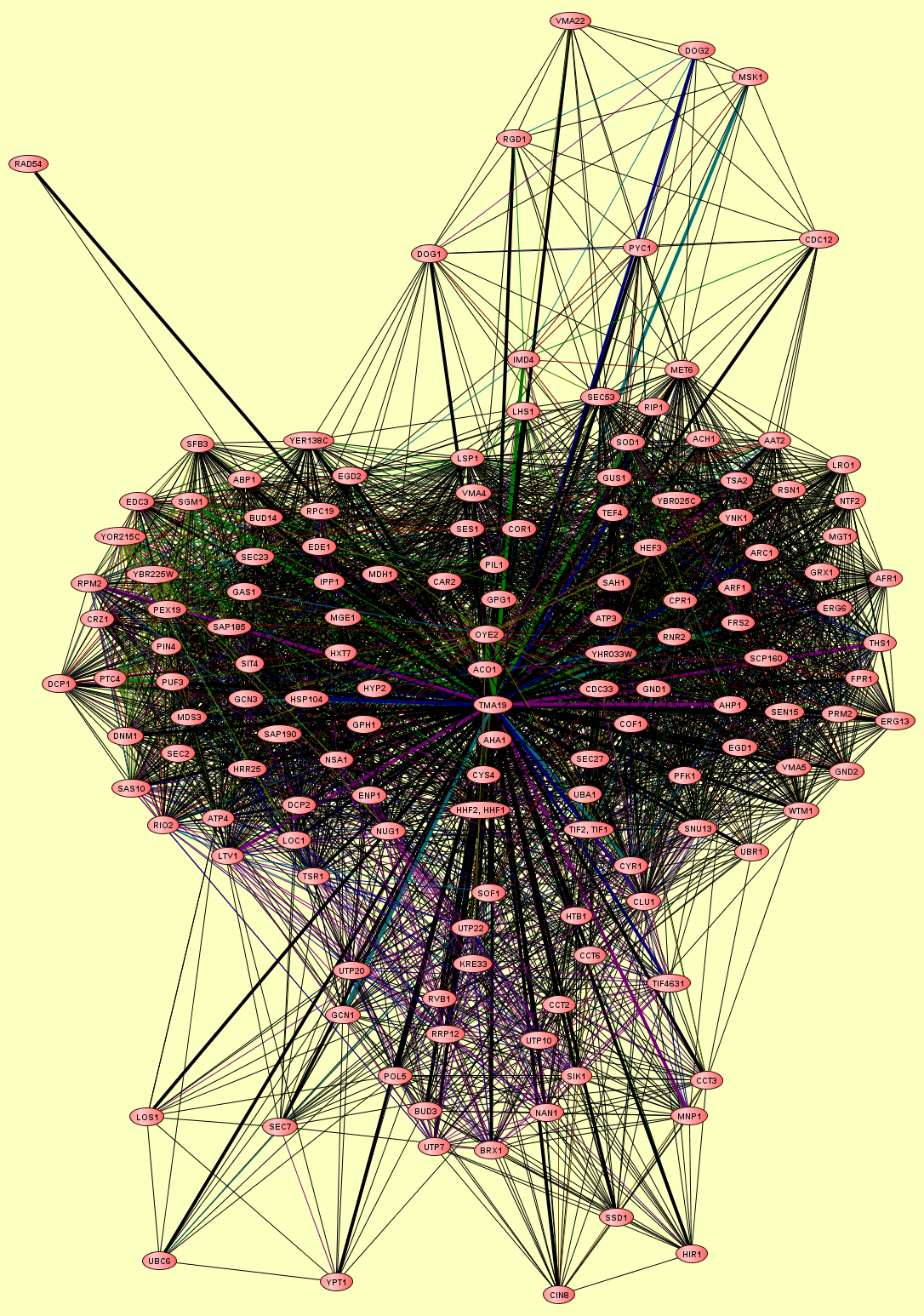
| LEGEND: |
|||
| = Same process, or one is unknown. | = Same branch, distance 4. | ||
| = Same branch, distance 1. | = Same branch, distance 5. | ||
| = Same branch, distance 2. | = Same branch, distance 6 or more. | ||
| = Same branch, distance 3. | = Not in same branch of GO. | ||
| Size | Notes | Members | |
| View GO Analysis | 11 proteins | This molecular complex record represents a population of complexes co-purified by immunoprecipitation of FLAG-tagged Ubc6. The topolgies of protein complexes in this experiment are unknown. | ATP4 GCN1 LOS1 POL5 RVB1 SEC7 TMA19 UBA1 UBC6 UTP20 YPT1 |
| View GO Analysis | 55 proteins | This molecular complex record represents a population of complexes co-purified by immunoprecipitation of FLAG-tagged Sen15. The topolgies of protein complexes in this experiment are unknown. | AAT2 ACH1 ACO1 AFR1 AHP1 ARC1 ARF1 ATP3 CAR2 CDC33 CLU1 COF1 COR1 CPR1 CYR1 CYS4 EGD1 ERG13 ERG6 FPR1 FRS2 GND1 GND2 GRX1 GUS1 HEF3 HHF1, HHF2 LRO1 LSP1 MET6 NTF2 OLA1 OYE2 PFK1 PIL1 PRM2 RNR2 RSN1 SAH1 SCP160 SEC53 SEN15 SES1 SNU13 SOD1 TEF4 THS1 TIF1, TIF2 TMA19 TSA2 UBA1 VMA4 VMA5 WTM1 YNK1 |
| View GO Analysis | 12 proteins | This molecular complex record represents a population of complexes co-purified by immunoprecipitation of FLAG-tagged Cdc12. The topolgies of protein complexes in this experiment are unknown. | CDC12 DOG1 DOG2 GUS1 IMD4 MET6 MSK1 PYC1 RGD1 SEC53 TMA19 VMA22 |
| View GO Analysis | 4 proteins | This molecular complex record represents a population of complexes co-purified by immunoprecipitation of FLAG-tagged Rad54. The topolgies of protein complexes in this experiment are unknown. | MDH1 MGE1 RAD54 TMA19 |
| View GO Analysis | 16 proteins | This molecular complex record represents a population of complexes co-purified by immunoprecipitation of FLAG-tagged Mgt1. The topolgies of protein complexes in this experiment are unknown. | AHA1 AHP1 ARF1 GND1 GPG1 HEF3 HTB1 LHS1 MGE1 MGT1 RIP1 SEC27 SOF1 TMA19 UBR1 YHR033W |
| View GO Analysis | 22 proteins | This molecular complex record represents a population of complexes co-purified by immunoprecipitation of FLAG-tagged Kre31. The topolgies of protein complexes in this experiment are unknown. | BRX1 BUD3 CCT2 CCT3 CCT6 CIN8 CLU1 HIR1 HTB1 KRE33 MNP1 NAN1 NOP56 POL5 RRP12 RVB1 SSD1 TIF4631 TMA19 UTP10 UTP22 UTP7 |
| View GO Analysis | 56 proteins | This molecular complex record represents a population of complexes co-purified by immunoprecipitation of FLAG-tagged Hrr25. The topolgies of protein complexes in this experiment are unknown. | ABP1 ACO1 AIM41 ATP4 BUD14 CAR2 COR1 CRZ1 CYS4 DCP1 DCP2 DNM1 EDC3 EDE1 EGD2 ENP1 GAS1 GCN3 GPH1 HHF1, HHF2 HRR25 HSP104 HXT7 HYP2 IPP1 LOC1 LSP1 LTV1 MDH1 MDS3 MGE1 NSA1 NUG1 OYE2 PEX19 PIL1 PIN4 PTC4 PUF3 RIO2 RPC19 RPM2 SAP185 SAP190 SAS10 SEC2 SEC23 SES1 SFB3 SGM1 SIT4 TMA19 TSR1 VMA4 YBR225W YER138C |