













| General Information: |
|
| Name(s) found: |
FCF2 /
YLR051C
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 217 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA nuclear outer membrane [IMP nucleolus [IDA |
| Biological Process: |
endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)
[IMP]
endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) [IMP] ribosome biogenesis [IEA] endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) [IMP] rRNA processing [IEA] |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDQSVEDLFG ALRDASASLE VKNSAKEQVS LQQEDVLQIG NNDDEVEIES KFQEIETNLK 60
61 KLPKLETGFD ALANKKKKKN VLPSVETEDK RKPNKSDKND NDWFTLPKPD DNMRREVQRD 120
121 LLLIKHRAAL DPKRHYKKQR WEVPERFAIG TIIEDKSEFY SSRMNRKERK STILETLMGD 180
181 EASNKYFKRK YNEIQEKSTS GRKAHYKKMK EMRKKRR |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Hazbun TR, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
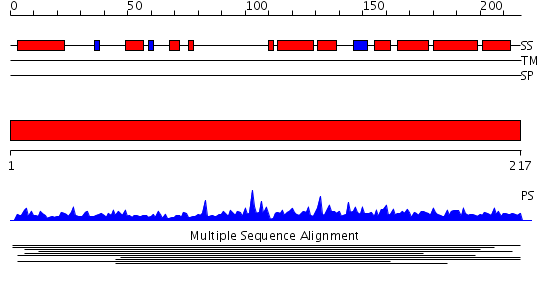
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..217] | 2.009966 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [86..217] | 48.259637 | No description for PF08698.2 was found. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)