













| General Information: |
|
| Name(s) found: |
EXO1 /
YOR033C
[SGD]
|
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 702 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IPI |
| Biological Process: |
DNA double-strand break processing
[IGI telomere maintenance [IGI mismatch repair [IMP |
| Molecular Function: |
5'-3' exonuclease activity
[IDA 5'-flap endonuclease activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGIQGLLPQL KPIQNPVSLR RYEGEVLAID GYAWLHRAAC SCAYELAMGK PTDKYLQFFI 60
61 KRFSLLKTFK VEPYLVFDGD AIPVKKSTES KRRDKRKENK AIAERLWACG EKKNAMDYFQ 120
121 KCVDITPEMA KCIICYCKLN GIRYIVAPFE ADSQMVYLEQ KNIVQGIISE DSDLLVFGCR 180
181 RLITKLNDYG ECLEICRDNF IKLPKKFPLG SLTNEEIITM VCLSGCDYTN GIPKVGLITA 240
241 MKLVRRFNTI ERIILSIQRE GKLMIPDTYI NEYEAAVLAF QFQRVFCPIR KKIVSLNEIP 300
301 LYLKDTESKR KRLYACIGFV IHRETQKKQI VHFDDDIDHH LHLKIAQGDL NPYDFHQPLA 360
361 NREHKLQLAS KSNIEFGKTN TTNSEAKVKP IESFFQKMTK LDHNPKVANN IHSLRQAEDK 420
421 LTMAIKRRKL SNANVVQETL KDTRSKFFNK PSMTVVENFK EKGDSIQDFK EDTNSQSLEE 480
481 PVSESQLSTQ IPSSFITTNL EDDDNLSEEV SEVVSDIEED RKNSEGKTIG NEIYNTDDDG 540
541 DGDTSEDYSE TAESRVPTSS TTSFPGSSQR SISGCTKVLQ KFRYSSSFSG VNANRQPLFP 600
601 RHVNQKSRGM VYVNQNRDDD CDDNDGKNQI TQRPSLRKSL IGARSQRIVI DMKSVDERKS 660
661 FNSSPILHEE SKKRDIETTK SSQARPAVRS ISLLSQFVYK GK |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
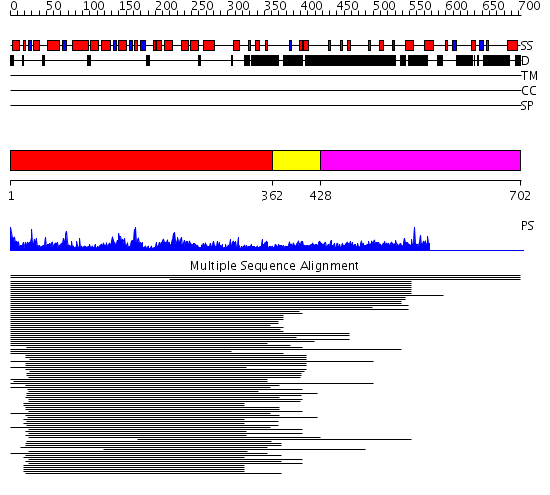
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..361] | 188.30103 | Flap endonuclease-1 (Fen-1 nuclease) | |
| 2 | View Details | [362..427] | 3.0 | Exonuclease domain of prokaryotic DNA polymerase; DNA polymerase I (Klenow fragment) | |
| 3 | View Details | [428..702] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)