













| General Information: |
|
| Name(s) found: |
MCR1 /
YKL150W
[SGD]
|
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 302 amino acids |
Gene Ontology: |
|
| Cellular Component: |
integral to mitochondrial outer membrane
[IDA mitochondrial intermembrane space [IDA mitochondrion [IDA mitochondrial outer membrane [IDA |
| Biological Process: |
response to oxidative stress
[IMP electron transport [IDA ergosterol biosynthetic process [IDA |
| Molecular Function: |
cytochrome-b5 reductase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFSRLSRSHS KALPIALGTV AIAAATAFYF ANRNQHSFVF NESNKVFKGD DKWIDLPISK 60
61 IEEESHDTRR FTFKLPTEDS EMGLVLASAL FAKFVTPKGS NVVRPYTPVS DLSQKGHFQL 120
121 VVKHYEGGKM TSHLFGLKPN DTVSFKGPIM KWKWQPNQFK SITLLGAGTG INPLYQLAHH 180
181 IVENPNDKTK VNLLYGNKTP QDILLRKELD ALKEKYPDKF NVTYFVDDKQ DDQDFDGEIS 240
241 FISKDFIQEH VPGPKESTHL FVCGPPPFMN AYSGEKKSPK DQGELIGILN NLGYSKDQVF 300
301 KF |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Ho Y, et al. (2002) |

|
ADK1 ATP3 ATP5 ATP7 CDC14 DPM1 FUR1 GLC7 HEF3 HMS1 MCR1 SNF4 SPE3 SSZ1 TPS1 TSA2 VAS1 |
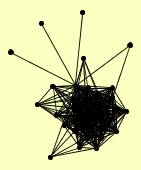
| View Details | Qiu et al. (2008) |
|
ALO1 ATP15 ATP2 ATP4 AYR1 CCP1 COB COR1 COX1 COX2 COX3 COX4 COX5A COX5B COX6 COX7 COX8 COX9 CYB2 CYC1 CYC7 CYT1 ERV1 MCR1 POR1 QCR10 QCR2 QCR7 QCR8 QCR9 RIP1 SDH2 SDH3 SDH4 TAZ1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | De Craene, J., et al. (2006) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
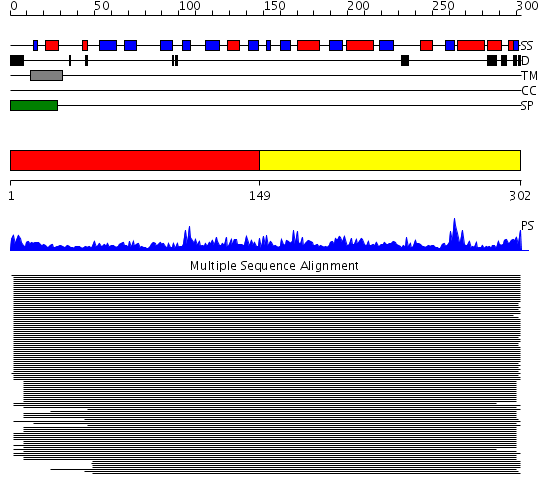
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..148] | 407.67837 | cytochrome b5 reductase | |
| 2 | View Details | [149..302] | 407.67837 | cytochrome b5 reductase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|