













| General Information: |
|
| Name(s) found: |
URA2 /
YJL130C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 2214 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA mitochondrion [IDA integral to membrane [IDA |
| Biological Process: |
'de novo' pyrimidine base biosynthetic process
[TAS glutamine metabolic process [IDA pyrimidine base biosynthetic process [IDA |
| Molecular Function: |
aspartate carbamoyltransferase activity
[IDA carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MATIAPTAPI TPPMESTGDR LVTLELKDGT VLQGYSFGAE KSVAGELVFQ TGMVGYPESV 60
61 TDPSYEGQIL VITYPLVGNY GVPDMHLRDE LVEELPRYFE SNRIHIAGLV ISHYTDEYSH 120
121 YLRKSSLGKW LQNEGIPAVY GVDTRSLTKH LRDAGSMLGR LSLEKSGSDR TISRSSSWRS 180
181 AFDVPEWVDP NVQNLVSKVS INEPKLYVPP ADNKHIELQT GPDGKVLRIL AIDVGMKYNQ 240
241 IRCFIKRGVE LKVVPWNYDF TKEDYDGLFI SNGPGDPSVL DDLSQRLSNV LEAKKTPVFG 300
301 ICLGHQLIAR AAGASTLKLK FGNRGHNIPC TSTISGRCYI TSQNHGFAVD VDTLTSGWKP 360
361 LFVNANDDSN EGIYHSELPY FSVQFHPEST PGPRDTEFLF DVFIQAVKEF KYTQVLKPIA 420
421 FPGGLLEDNV KAHPRIEAKK VLVLGSGGLS IGQAGEFDYS GSQAIKALKE EGIYTILINP 480
481 NIATIQTSKG LADKVYFVPV TAEFVRKVIL HERPDAIYVT FGGQTALSVG IAMKDEFEAL 540
541 GVKVLGTPID TIITTEDREL FSNAIDEINE KCAKSQAANS VDEALAAVKE IGFPVIVRAA 600
601 YALGGLGSGF ANNEKELVDL CNVAFSSSPQ VLVEKSMKGW KEVEYEVVRD AFDNCITVCN 660
661 MENFDPLGIH TGDSIVVAPS QTLSDEDYNM LRTTAVNVIR HLGVVGECNI QYALNPVSKD 720
721 YCIIEVNARL SRSSALASKA TGYPLAYTAA KLGLNIPLNE VKNSVTKSTC ACFEPSLDYC 780
781 VVKMPRWDLK KFTRVSTELS SSMKSVGEVM SIGRTFEEAI QKAIRSTEYA NLGFNETDLD 840
841 IDIDYELNNP TDMRVFAIAN AFAKKGYSVD KVWEMTRIDK WFLNKLHDLV QFAEKISSFG 900
901 TKEELPSLVL RQAKQLGFDD RQIARFLDSN EVAIRRLRKE YGITPFVKQI DTVAAEFPAY 960
961 TNYLYMTYNA DSHDLSFDDH GVMVLGSGVY RIGSSVEFDW CAVTAVRTLR ANNIKTIMVN1020
1021 YNPETVSTDY DEADRLYFET INLERVLDIY EIENSSGVVV SMGGQTSNNI AMTLHRENVK1080
1081 ILGTSPDMID SAENRYKFSR MLDQIGVDQP AWKELTSMDE AESFAEKVGY PVLVRPSYVL1140
1141 SGAAMNTVYS KNDLESYLNQ AVEVSRDYPV VITKYIENAK EIEMDAVARN GELVMHVVSE1200
1201 HVENAGVHSG DATLIVPPQD LAPETVDRIV VATAKIGKAL KITGPYNIQF IAKDNEIKVI1260
1261 ECNVRASRSF PFISKVVGVN LIELATKAIM GLPLTPYPVE KLPDDYVAVK VPQFSFPRLA1320
1321 GADPVLGVEM ASTGEVATFG HSKYEAYLKS LLATGFKLPK KNILLSIGSY KEKQELLSSV1380
1381 QKLYNMGYKL FATSGTADFL SEHGIAVQYL EVLNKDDDDQ KSEYSLTQHL ANNEIDLYIN1440
1441 LPSANRFRRP ASYVSKGYKT RRLAVDYSVP LVTNVKCAKL LIEAISRNIT LDVSERDAQT1500
1501 SHRTITLPGL INIATYVPNA SHVIKGPAEL KETTRLFLES GFTYCQLMPR SISGPVITDV1560
1561 ASLKAANSVS QDSSYTDFSF TIAGTAHNAH SVTQSASKVT ALFLPLRELK NKITAVAELL1620
1621 NQWPTEKQVI AEAKTADLAS VLLLTSLQNR SIHITGVSNK EDLALIMTVK AKDPRVTCDV1680
1681 NIYSLFIAQD DYPEAVFLPT KEDQEFFWNN LDSIDAFSVG ALPVALANVT GNKVDVGMGI1740
1741 KDSLPLLLAA VEEGKLTIDD IVLRLHDNPA KIFNIPTQDS VVEIDLDYSF RRNKRWSPFN1800
1801 KDMNGGIERV VYNGETLVLS GELVSPGAKG KCIVNPSPAS ITASAELQST SAKRRFSITE1860
1861 EAIADNLDAA EDAIPEQPLE QKLMSSRPPR ELVAPGAIQN LIRSNNPFRG RHILSIKQFK1920
1921 RSDFHVLFAV AQELRAAVAR EGVLDLMKGH VITTIFFEPS TRTCSSFIAA MERLGGRIVN1980
1981 VNPLVSSVKK GETLQDTIRT LACYSDAIVM RHSEEMSVHI AAKYSPVPII NGGNGSREHP2040
2041 TQAFLDLFTI REEIGTVNGI TVTFMGDLKH GRTVHSLCRL LMHYQVRINL VSPPELRLPE2100
2101 GLREELRKAG LLGVESIELT PHIISKTDVL YCTRVQEERF NSPEEYARLK DTYIVDNKIL2160
2161 AHAKENMAIM HPLPRVNEIK EEVDYDHRAA YFRQMKYGLF VRMALLAMVM GVDM |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
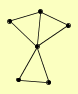
|
NPL3 RPS0B RPS4B, RPS4A URA2 VMA2 YDJ1 |
| View Details | Krogan NJ, et al. (2006) |
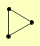
|
CPA1 CPA2 URA2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Cheeseman IM, et al. (2002) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | 3912: TAP-tagged, strain background includes hpc2(delta) | Green EM, et al (2005) | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | phosphorylation data - cascade search | Green EM, et al (2005) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi 1 with ha tag from october 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi1 with gst from october 2005 | McCusker D, et al (2007) | |
| View Run | Sample bob1 (2nd set) from october 2005 | McCusker D, et al (2007) | |
| View Run | Sample: HX11 - both fusion and trans-SNARE complex formation take place. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | Sample: HX14 - mixture in detergent (control). | Hao Xu, et al. (2010) | |
| View Run | hx23: mixture in detergent (control) Investigating Vam3p, Nyv1p and their partners in trans-SNARE complex | Hao Xu, et al. (2010) | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #13 Mitotic Prep2-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #02 Alpha Factor Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #08 Alpha Factor Prep4-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
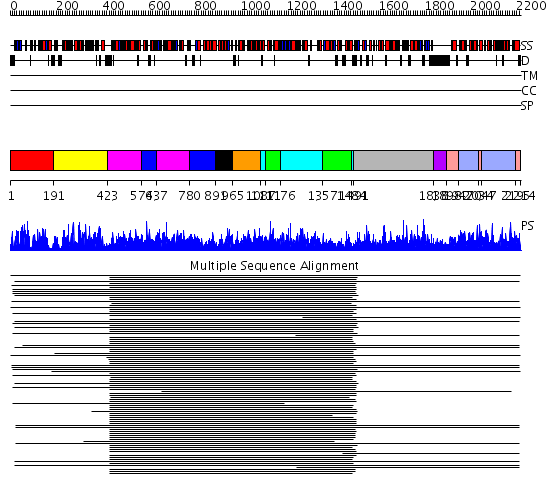
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..190] | 764.0 | Carbamoyl phosphate synthetase, small subunit N-terminal domain; Carbamoyl phosphate synthetase, small subunit C-terminal domain | |
| 2 | View Details | [191..422] | 764.0 | Carbamoyl phosphate synthetase, small subunit N-terminal domain; Carbamoyl phosphate synthetase, small subunit C-terminal domain | |
| 3 | View Details | [423..573] [637..779] |
11000.0 | Carbamoyl phosphate synthetase, large subunit connection domain; Carbamoyl phosphate synthetase, large subunit allosteric, C-terminal domain; Carbamoyl phosphate synthetase (CPS), large subunit PreATP-grasp domains; Carbamoyl phosphate synthetase (CPS), large subunit ATP-binding domains | |
| 4 | View Details | [574..636] [780..890] |
11000.0 | Carbamoyl phosphate synthetase, large subunit connection domain; Carbamoyl phosphate synthetase, large subunit allosteric, C-terminal domain; Carbamoyl phosphate synthetase (CPS), large subunit PreATP-grasp domains; Carbamoyl phosphate synthetase (CPS), large subunit ATP-binding domains | |
| 5 | View Details | [891..964] | 11000.0 | Carbamoyl phosphate synthetase, large subunit connection domain; Carbamoyl phosphate synthetase, large subunit allosteric, C-terminal domain; Carbamoyl phosphate synthetase (CPS), large subunit PreATP-grasp domains; Carbamoyl phosphate synthetase (CPS), large subunit ATP-binding domains | |
| 6 | View Details | [965..1086] | 11000.0 | Carbamoyl phosphate synthetase, large subunit connection domain; Carbamoyl phosphate synthetase, large subunit allosteric, C-terminal domain; Carbamoyl phosphate synthetase (CPS), large subunit PreATP-grasp domains; Carbamoyl phosphate synthetase (CPS), large subunit ATP-binding domains | |
| 7 | View Details | [1087..1110] [1176..1356] [1484..1490] |
11000.0 | Carbamoyl phosphate synthetase, large subunit connection domain; Carbamoyl phosphate synthetase, large subunit allosteric, C-terminal domain; Carbamoyl phosphate synthetase (CPS), large subunit PreATP-grasp domains; Carbamoyl phosphate synthetase (CPS), large subunit ATP-binding domains | |
| 8 | View Details | [1111..1175] [1357..1483] |
11000.0 | Carbamoyl phosphate synthetase, large subunit connection domain; Carbamoyl phosphate synthetase, large subunit allosteric, C-terminal domain; Carbamoyl phosphate synthetase (CPS), large subunit PreATP-grasp domains; Carbamoyl phosphate synthetase (CPS), large subunit ATP-binding domains | |
| 9 | View Details | [1491..1837] | 32.0 | L-hydantoinase | |
| 10 | View Details | [1838..1897] | N/A | No confident structure predictions are available. | |
| 11 | View Details | [1898..1946] [2034..2046] [2195..2214] |
717.9691 | Aspartate carbamoyltransferase catalytic subunit | |
| 12 | View Details | [1947..2033] [2047..2194] |
717.9691 | Aspartate carbamoyltransferase catalytic subunit |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 10 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 11 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 12 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)