













| General Information: |
|
| Name(s) found: |
PFK2 /
YMR205C
[SGD]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 959 amino acids |
Gene Ontology: |
|
| Cellular Component: |
6-phosphofructokinase complex
[IMP cytoplasm [IDA mitochondrion [IDA |
| Biological Process: |
glycolysis
[IDA |
| Molecular Function: |
6-phosphofructokinase activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTVTTPFVNG TSYCTVTAYS VQSYKAAIDF YTKFLSLENR SSPDENSTLL SNDSISLKIL 60
61 LRPDEKINKN VEAHLKELNS ITKTQDWRSH ATQSLVFNTS DILAVKDTLN AMNAPLQGYP 120
121 TELFPMQLYT LDPLGNVVGV TSTKNAVSTK PTPPPAPEAS AESGLSSKVH SYTDLAYRMK 180
181 TTDTYPSLPK PLNRPQKAIA VMTSGGDAPG MNSNVRAIVR SAIFKGCRAF VVMEGYEGLV 240
241 RGGPEYIKEF HWEDVRGWSA EGGTNIGTAR CMEFKKREGR LLGAQHLIEA GVDALIVCGG 300
301 DGSLTGADLF RSEWPSLIEE LLKTNRISNE QYERMKHLNI CGTVGSIDND MSTTDATIGA 360
361 YSALDRICKA IDYVEATANS HSRAFVVEVM GRNCGWLALL AGIATSADYI FIPEKPATSS 420
421 EWQDQMCDIV SKHRSRGKRT TIVVVAEGAI AADLTPISPS DVHKVLVDRL GLDTRITTLG 480
481 HVQRGGTAVA YDRILATLQG LEAVNAVLES TPDTPSPLIA VNENKIVRKP LMESVKLTKA 540
541 VAEAIQAKDF KRAMSLRDTE FIEHLNNFMA INSADHNEPK LPKDKRLKIA IVNVGAPAGG 600
601 INSAVYSMAT YCMSQGHRPY AIYNGWSGLA RHESVRSLNW KDMLGWQSRG GSEIGTNRVT 660
661 PEEADLGMIA YYFQKYEFDG LIIVGGFEAF ESLHQLERAR ESYPAFRIPM VLIPATLSNN 720
721 VPGTEYSLGS DTALNALMEY CDVVKQSASS TRGRAFVVDC QGGNSGYLAT YASLAVGAQV 780
781 SYVPEEGISL EQLSEDIEYL AQSFEKAEGR GRFGKLILKS TNASKALSAT KLAEVITAEA 840
841 DGRFDAKPAY PGHVQQGGLP SPIDRTRATR MAIKAVGFIK DNQAAIAEAR AAEENFNADD 900
901 KTISDTAAVV GVKGSHVVYN SIRQLYDYET EVSMRMPKVI HWQATRLIAD HLVGRKRVD |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
PFK1 PFK2 |
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
PFK1 PFK2 |
| View Details | Krogan NJ, et al. (2006) |

|
PFK1 PFK2 |
| View Details | Gavin AC, et al. (2006) |

|
PFK1 PFK2 RPS10A RPS22A |
| View Details | Gavin AC, et al. (2002) |

|
ADA2 ADH1 CLU1 FUN19 GCN1 GCN5 HEM13 ISW1 KAP114 KAP123 MED4 MES1 MNN4 MOT1 MYO1 NGG1 NOP1 PDC1 PFK1 PFK2 PSK1 PTC1 PUF6 RPB2 RPN9 RPO21 RPT5 RVB2 SGF29 SGF73 SIN3 SIN4 SPT15 SPT20 SPT7 SPT8 TAF1 TAF10 TAF11 TAF12 TAF13 TAF14 TAF2 TAF3 TAF4 TAF5 TAF6 TAF7 TAF8 TAF9 TBF1 TEF4 VAS1 VPS1 YEF3 |
| View Details | Ho Y, et al. (2002) |

|
ACS2 ADE3 ADE6 ADO1 ALA1 APE2 ARA1 BAT1 CBF5 CDC10 CLU1 CYS3 DED81 DPS1 ERG10 ERG20 FPR1 GCY1 GPH1 GRS1 HOM2 HOM3 HYP2 ILS1 IMD3 KRS1 LSP1 MDH3 MLP2 OLA1 PAB1 PFK1 PFK2 PGM2 PMI40 PRP6 RNR2 SAC6 SAP155 SCP160 SHQ1 SIT4 SOD1 SRP1 SSZ1 TAP42 THR4 THS1 TIF1, TIF2 TRR1 UBA1 URA1 YDL124W YDR341C |
| View Details | Qiu et al. (2008) |
|
GLK1 HXK2 PFK1 PFK2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Toshima J, et al (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi 1 with ha tag from october 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi1 with gst from october 2005 | McCusker D, et al (2007) | |
| View Run | Sample bob1 (2nd set) from october 2005 | McCusker D, et al (2007) | |
| View Run | #19 Asynchronous Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #35 Asynchronous SPB prep | Keck JM, et al. (2011) | |
| View Run | #04 Alpha Factor Prep2 | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #19b Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) | |
| View Run | #19a Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
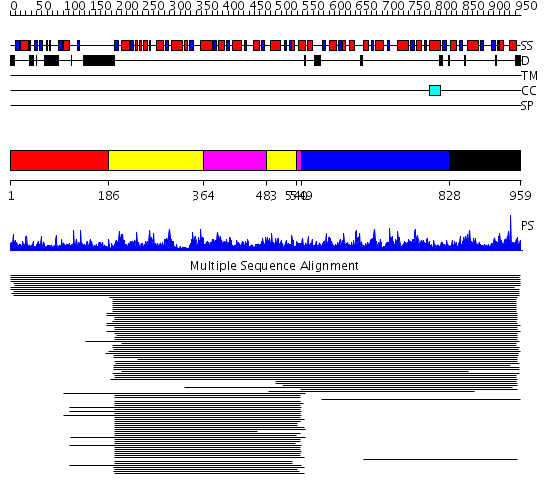
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..185] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [186..363] [483..539] |
635.9897 | ATP-dependent phosphofructokinase | |
| 3 | View Details | [364..482] [540..548] |
635.9897 | ATP-dependent phosphofructokinase | |
| 4 | View Details | [549..827] | 112.08661 | Pyrophosphate-dependent phosphofructokinase | |
| 5 | View Details | [828..959] | 12.5 | ATP-dependent phosphofructokinase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)