













| General Information: |
|
| Name(s) found: |
URB2 /
YJR041C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1174 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleolus
[IDA |
| Biological Process: |
rRNA metabolic process
[IMP ribosome biogenesis [IPI |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGDLTEELSI PDNAQDLSKL LRSTSTKPHQ IAEIVSKFDK LETYFPKKEI FVLDLLIDRL 60
61 NNGNLDDFKT SEHTWIIFTR LLDAINDPIS IKKLLKKLKT VPVMIRTFFL WPKDKLLTRS 120
121 VSFIKAFFAI NDYLIVNFSV EESFQLLEHA INGLSSCPTT DFALSYLQDA CNLTHVDNIT 180
181 TTDNKIATCY CKHMLLPSLR YFAQTKNSAS SNQSFIRLSH FMGKFLLQPR IDYMKLNKKF 240
241 VQENASEITD DMAYYYFATF VTFLSKDNFA QLEVIFTILG AKKPSLECRF LNLLSESKKT 300
301 VSQEFLEALL LEMLASTDES GVLSLIPIIL KLDIEVAIKH IFRLLELIQL ENLNDPLFSS 360
361 HIWDLIIQSH ANARELSDFF AKINEYCSRK GPDSYFLINH PAYVKSITKQ LFTLSSLQWK 420
421 NLLQALLDQV NHDSTNRVPL YLIRICLEGL SEGASRATLD EVKPILSQVF TLESFNNSLQ 480
481 WDLKYHIMEV YDDIVPAEEL EKIDYVLSSN IFDTTSADVE ELFFYCFKLR EYISFDLSDA 540
541 KKKFMRHFEI LDEERKSNLS YSVVSKFATL VNNNFTREQI SSLIDSLLLN STNLSSLLKN 600
601 DDIFEETNIT YALINKLASS YHQTFALEAL IQIPIQCINK NVRVALINNL TCESFCLDSA 660
661 TRECLLHLLS SPTFKSNIET NFYELCEKTI MSPEMAISET GDEKKEIEDK ISIFEKVWTN 720
721 HLSQAKEPVS EKFLESGYDI VKQSMSLSNG DSKLIIAGFT IAKFLKPDNK HRDIQGMAIS 780
781 YAVKILENYS ENFESETIPL FRISMSTLYK IITTGQGDIS KHKSRILDIF SKIMLRYHSK 840
841 KVYHAPEEQE MFLVHSLLTE NKLEYIFAEY LNIEHTDKCD SALGFCLEES LKQGPDAFNR 900
901 LLWNSAKSFS TISQPCAEKF VRVFIIMSKR IARDNNLGHH LFVIALLEAY TYCDIEKFGY 960
961 KSYLLLFNAI KEFLVSKPWL FSQYCIEMLL PFCLKTLAFI VNHESTDEIN EGFINIIEVI1020
1021 DHMLLVHRFK FSNRHHLFNS VLCQILEIIA IHDGTLCANS ADAVARLITN YCEPYNVSNA1080
1081 QNGQKNNLSS KISLIKQSIR KNVLVVLTKY IQLSITTQFS LNIKKSLQPG IHAIFDILSQ1140
1141 NELNQLNAFL DTPGKQYFKA LYLQYKKVGK WRED |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
URB1 URB2 |
| View Details | Krogan NJ, et al. (2006) |

|
CBC2 KAP95 NUP1 NUP2 NUP60 OGG1 PCT1 RAD53 SRP1 STO1 ULP1 URB2 YRA2 |
| View Details | Gavin AC, et al. (2006) |
|
NOP8 URB1 URB2 |
| View Details | Ho Y, et al. (2002) |

|
CBF5 CBP2 CCM1 CDC33 CIC1 DBP7 DRS1 ENP2 ERB1 GBP2 HAS1 HMO1 HTB2 IMD1 IMD3 ISA1 KRE33 KRI1 KRR1 MGM101 MSS116 NOC2 NOP12 NOP6 NPL3 PET127 POL5 PRP43 PUF6 PWP1 RLI1 RPF2 RRP12 RRP5 TIF4631 TIF6 TRA1 TSR1 URB1 URB2 UTP10 UTP20 YPP1 YRA1 YTM1 |
| View Details | Qiu et al. (2008) |
|
DHR2 GAR1 MAK5 URB2 UTP6 UTP8 UTP9 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Cheeseman IM, et al. (2002) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
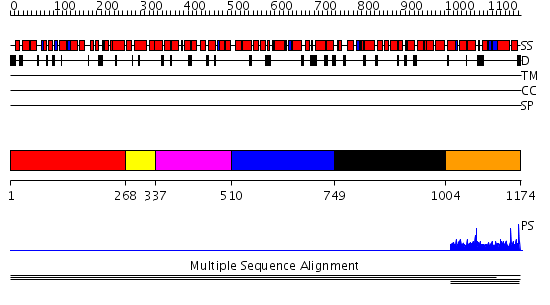
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..267] | 1.6 | Constant regulatory domain of protein phosphatase 2a, pr65alpha | |
| 2 | View Details | [268..336] | 1.6 | Constant regulatory domain of protein phosphatase 2a, pr65alpha | |
| 3 | View Details | [337..509] | 1.6 | Constant regulatory domain of protein phosphatase 2a, pr65alpha | |
| 4 | View Details | [510..748] | 1.6 | Constant regulatory domain of protein phosphatase 2a, pr65alpha | |
| 5 | View Details | [749..1003] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [1004..1174] | 1.003931 | View MSA. Confident ab initio structure predictions are available. | |
| 7 | View Details | [767..977] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [978..1174] | 4.016994 | View MSA. Confident ab initio structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)