













| General Information: |
|
| Name(s) found: |
MNN1 /
YER001W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 762 amino acids |
Gene Ontology: |
|
| Cellular Component: |
Golgi apparatus
[IDA |
| Biological Process: |
protein amino acid O-linked glycosylation
[IDA N-glycan processing [IMP |
| Molecular Function: |
alpha-1,3-mannosyltransferase activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLALRRFILN QRSLRSCTIP ILVGALIIIL VLFQLVTHRN DALIRSSNVN STNKKTLKDA 60
61 DPKVLIEAFG SPEVDPVDTI PVSPLELVPF YDQSIDTKRS SSWLINKKGY YKHFNELSLT 120
121 DRCKFYFRTL YTLDDEWTNS VKKLEYSIND NEGVDEGKDA NGNPMDEKSE RLYRRKYDMF 180
181 QAFERIRAYD RCFMQANPVN IQEIFPKSDK MSKERVQSKL IKTLNATFPN YDPDNFKKYD 240
241 QFEFEHKMFP FINNFTTETF HEMVPKITSP FGKVLEQGFL PKFDHKTGKV QEYFKYEYDP 300
301 SKTFWANWRD MSAKVAGRGI VLSLGSNQFP LAVKFIASLR FEGNTLPIQV VYRGDELSQE 360
361 LVDKLIYAAR SPDFKPVENN YDNSTNVPQE IWFLDVSNTI HPKWRGDFGS YKSKWLVVLL 420
421 NLLQEFVFLD IDAISYEKID NYFKTTEYQK TGTVFYRERA LRENVNERCI ARYETLLPRN 480
481 LESKNFQNSL LIDPDHALNE CDNTLTTEEY IFKAFFHHRR QHQLEAGLFA VDKSKHTIPL 540
541 VLAAMIHLAK NTAHCTHGDK ENFWLGFLAA GHTYALQGVY SGAIGDYVKK TDLNGKRQEA 600
601 AVEICSGQIA HMSTDKKTLL WVNGGGTFCK HDNAAKDDWK KDGDFKKFKD QFKTFEEMEK 660
661 YYYITPISSK YVILPDPKSD DWHRASAGAC GGYIWCATHK TLLKPYSYNH RTTHGELITL 720
721 DEEQRLHIDA VNTVWSHANK DNTRSFTEEE IKELENSRHE QS |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
KTI11 MNN1 |
| View Details | Krogan NJ, et al. (2006) |
|
AIM22 DPH1 DPH2 EFT2, EFT1 HGH1 MNN1 PGI1 RRG10 TRX1 YMR315W |
| View Details | Qiu et al. (2008) |
|
ANP1 GNT1 KRE2 KTR1 KTR4 MNN1 MNN11 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | 3829 - low filter | Green EM, et al (2005) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | sample: hx3 | Hao Xu, et al. (2010) | |
| View Run | Sample: HX11 - both fusion and trans-SNARE complex formation take place. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | Sample: HX14 - mixture in detergent (control). | Hao Xu, et al. (2010) | |
| View Run | hx23: mixture in detergent (control) Investigating Vam3p, Nyv1p and their partners in trans-SNARE complex | Hao Xu, et al. (2010) | |
| View Run | #20 Asynchronous Prep1-TiO2 Phosphopeptide enrichment, PartB | Keck JM, et al. (2011) | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
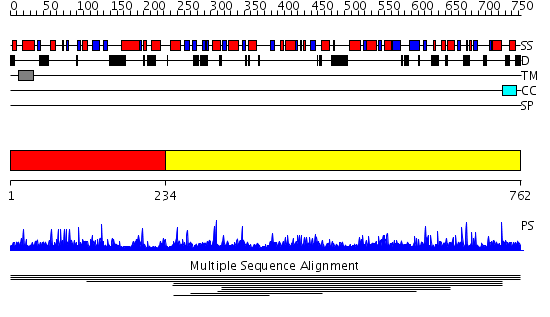
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..233] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [234..762] | 2.008843 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [302..762] | 1.51 | apo form of the 162S mutant of glycogenin |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|