













| General Information: |
|
| Name(s) found: |
KTR4 /
YBR199W
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 464 amino acids |
Gene Ontology: |
|
| Cellular Component: |
Golgi apparatus
[ISS |
| Biological Process: |
protein amino acid N-linked glycosylation
[ISS |
| Molecular Function: |
mannosyltransferase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRFLSKRILK PVLSVIILIS IAVTVVLYFL TANENYLQAV KDSAKSQYAS LRESYKSITG 60
61 KTESADELPD HDAEVLDSIM DRLHEPLYEK DTFDPNEVLA ENKQLYEEFL LQEISEPKVD 120
121 NLVRSGDPLA GKAKGTILSL VRNSDLEDII SSIQQLEEEY NKNFGYPYTF LNDEEFTDEF 180
181 KDGIKSILPK DRVVEFGTIG PDNWNMPDSI DRERYDQEMD KMSKENIQYA EVESYHNMCR 240
241 FYSKEFYHHP LLSKYKYVWR LEPNVNFYCK INYDVFQFMN KNDKIYGFVL NLYDSPQTIE 300
301 TLWTSTMDFV EEHPNYLNVN GAFAWLKDNS QNPKNYDYTQ GYSTCHFWTN FEIVDLDFLR 360
361 SEPYEKYMQY LEEKGGFYYE RWGDAPVRSL ALALFADKSS IHWFRDIGYH HTPYTNCPTC 420
421 PADSDRCNGN CVPGKFTPWS DLDNQNCQAT WIRHSMSEEE LEMY |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Qiu et al. (2008) |
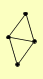
|
GNT1 KRE2 KTR4 MNN1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Sample: HX13 - trans-SNARE complex forms but fusion is blocked. | Hao Xu, et al. (2010) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
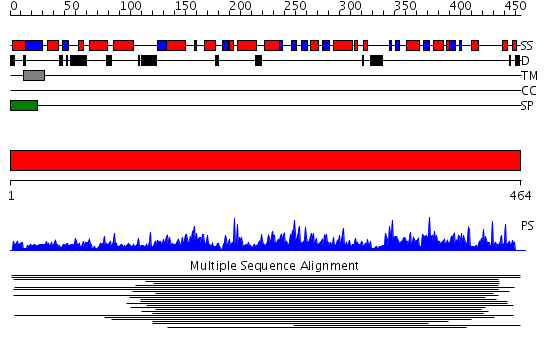
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..464] | 222.142668 | Glycolipid 2-alpha-mannosyltransferase Confident ab initio structure predictions are available. | |
| 2 | View Details | [116..464] | 148.0 | Crystal structure of yeast alpha1,2-mannosyltransferase Kre2p/Mnt1p |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|