













| General Information: |
|
| Name(s) found: |
RAD52 /
YML032C
[SGD]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 471 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA nuclear chromosome [IDA |
| Biological Process: |
meiotic DNA recombinase assembly
[TAS double-strand break repair via break-induced replication [TAS double-strand break repair via single-strand annealing [IGI telomere maintenance via recombination [IMP double-strand break repair via synthesis-dependent strand annealing [TAS DNA recombinase assembly [IDA postreplication repair [IMP |
| Molecular Function: |
DNA strand annealing activity
[IDA recombinase activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNEIMDMDEK KPVFGNHSED IQTKLDKKLG PEYISKRVGF GTSRIAYIEG WRVINLANQI 60
61 FGYNGWSTEV KSVVIDFLDE RQGKFSIGCT AIVRVTLTSG TYREDIGYGT VENERRKPAA 120
121 FERAKKSAVT DALKRSLRGF GNALGNCLYD KDFLAKIDKV KFDPPDFDEN NLFRPTDEIS 180
181 ESSRTNTLHE NQEQQQYPNK RRQLTKVTNT NPDSTKNLVK IENTVSRGTP MMAAPAEANS 240
241 KNSSNKDTDL KSLDASKQDQ DDLLDDSLMF SDDFQDDDLI NMGNTNSNVL TTEKDPVVAK 300
301 QSPTASSNPE AEQITFVTAK AATSVQNERY IGEESIFDPK YQAQSIRHTV DQTTSKHIPA 360
361 SVLKDKTMTT ARDSVYEKFA PKGKQLSMKN NDKELGPHML EGAGNQVPRE TTPIKTNATA 420
421 FPPAAAPRFA PPSKVVHPNG NGAVPAVPQQ RSTRREVGRP KINPLHARKP T |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
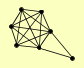
|
MPH1 RAD51 RAD52 RFA1 RFA2 RFA3 RIM1 |
| View Details | (MIPS) Mewes HW, et al. (2004) |
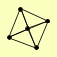
|
RAD51 RAD52 RAD54 RAD55 RAD57 |
| View Details | Krogan NJ, et al. (2006) |

|
RAD51 RAD52 |
| View Details | Gavin AC, et al. (2006) |
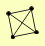
|
MPH1 MSH6 RAD52 RFA1 |
| View Details | Gavin AC, et al. (2002) |

|
ARO1 CCT8 CLU1 CMR1 CPA2 DNA2 ECM29 GCN20 GFA1 GUS1 HDA1 HDA2 HDA3 HSC82 IDH2 ILV1 KAP123 KAP95 LCD1 MEC1 MGM101 MKT1 MLH1 MPH1 MRPS28 MSH2 MSH3 MSH6 OLA1 PFK1 PGK1 PMS1 PRO2 RAD16 RAD23 RAD34 RAD4 RAD51 RAD52 RFA1 RFA2 RFA3 RIM1 RNH1 RPA135 RPB2 RPN10 RPN12 RPN3 RPN6 RPN8 RPN9 RPT2 RPT3 RPT6 RSC8 RVB2 SGS1 SHS1 SRO7 SSL1 TFA1 TFB1 TFB4 TIF1, TIF2 TOP3 VAS1 VPS1 YHB1 YHR122W |
| View Details | Ho Y, et al. (2002) |

|
AAC3 AHA1 ALD5 ATP3 BEM2 ECM10 GCD11 HOM3 HOR2 ILV2 OYE2 PIL1 PRB1 PTC3 RAD52 RAD59 RHR2 SEC27 SEC53 TEF4 UBA1 VMA8 YER138C YPT31 |
| View Details | Qiu et al. (2008) |
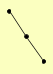
|
RAD18 RAD51 RAD52 |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
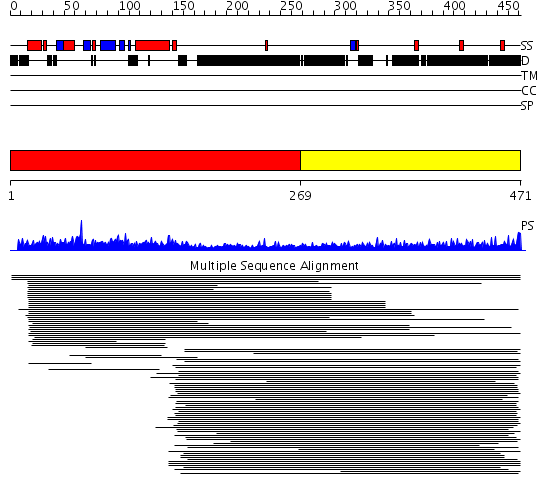
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..268] | 75.69897 | The homologous-pairing domain of Rad52 recombinase | |
| 2 | View Details | [269..471] | 8.192997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)