













| General Information: |
|
| Name(s) found: |
SPT5 /
YML010W
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1063 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA transcription elongation factor complex [IPI DSIF complex [IPI mitochondrion [IDA |
| Biological Process: |
regulation of transcription, DNA-dependent
[IGI chromatin organization [TAS RNA elongation from RNA polymerase II promoter [IPI |
| Molecular Function: |
RNA polymerase II transcription elongation factor activity
[IGI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSDNSDTNVS MQDHDQQFAD PVVVPQSTDT KDENTSDKDT VDSGNVTTTE STERAESTSN 60
61 IPPLDGEEKE AKSEPQQPED NAETAATEQV SSSNGPATDD AQATLNTDSS EANEIVKKEE 120
121 GSDERKRPRE EDTKNSDGDT KDEGDNKDED DDEDDDDDDD DEDDDDEAPT KRRRQERNRF 180
181 LDIEAEVSDD EDEDEDEEDS ELVREGFITH GDDEDDEASA PGARRDDRLH RQLDQDLNKT 240
241 SEEDAQRLAK ELRERYGRSS SKQYRAAAQD GYVPQRFLLP SVDTATIWGV RCRPGKEKEL 300
301 IRKLLKKKFN LDRAMGKKKL KILSIFQRDN YTGRIYIEAP KQSVIEKFCN GVPDIYISQK 360
361 LLIPVQELPL LLKPNKSDDV ALEEGSYVRI KRGIYKGDLA MVDQISENNL EVMLKIVPRL 420
421 DYGKFDEIDP TTQQRKSRRP TFAHRAPPQL FNPTMALRLD QANLYKRDDR HFTYKNEDYI 480
481 DGYLYKSFRI QHVETKNIQP TVEELARFGS KEGAVDLTSV SQSIKKAQAA KVTFQPGDRI 540
541 EVLNGEQRGS KGIVTRTTKD IATIKLNGFT TPLEFPISTL RKIFEPGDHV TVINGEHQGD 600
601 AGLVLMVEQG QVTFMSTQTS REVTITANNL SKSIDTTATS SEYALHDIVE LSAKNVACII 660
661 QAGHDIFKVI DETGKVSTIT KGSILSKINT ARARVSSVDA NGNEIKIGDT IVEKVGSRRE 720
721 GQVLYIQTQQ IFVVSKKIVE NAGVFVVNPS NVEAVASKDN MLSNKMDLSK MNPEIISKMG 780
781 PPSSKTFQQP IQSRGGREVA LGKTVRIRSA GYKGQLGIVK DVNGDKATVE LHSKNKHITI 840
841 DKHKLTYYNR EGGEGITYDE LVNRRGRVPQ ARMGPSYVSA PRNMATGGIA AGAAATSSGL 900
901 SGGMTPGWSS FDGGKTPAVN AHGGSGGGGV SSWGGASTWG GQGNGGASAW GGAGGGASAW 960
961 GGQGTGATST WGGASAWGNK SSWGGASTWA SGGESNGAMS TWGGTGDRSA YGGASTWGGN1020
1021 NNNKSTRDGG ASAWGNQDDG NRSAWNNQGN KSNYGGNSTW GGH |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
DST1 IWR1 PPH3 PSY2 PSY4 RPB10 RPB11 RPB2 RPB3 RPB4 RPB5 RPB7 RPB8 RPB9 RPO21 RPO26 SPT4 SPT5 SSD1 TFG1 TFG2 |
| View Details | Krogan NJ, et al. (2006) |
|
ABF2 ARG82 CKI1 CMR1 HHF1, HHF2 HTA1 HTA2 HTB1 HTB2 NUP188 PPH3 PSY2 PSY4 RRD1 SLX5 SLX8 SPT5 STP1 TOP2 |
| View Details | Gavin AC, et al. (2002) |
|
ABD1 AOS1 ASC1 ATG9 BEM2 CLU1 FUN12 GLT1 HCR1 IST2 IWR1 KTR5 LSG1 NIP1 NOG1 NOP7 PPH3 PRT1 PSY2 PSY4 RGA2 RIC1 RLI1 RPB11 RPB2 RPB3 RPB4 RPB5 RPB7 RPB8 RPB9 RPG1 RPO21 RPO26 SPT4 SPT5 SUA7 SUI1 SUI2 SUI3 SUP45 TAF14 TDH3 TFG1 TFG2 TIF34 TIF35 TIF5 TRM3 UTP6 YGL036W |
| View Details | Ho Y, et al. (2002) |

|
BCY1 CAR2 CPR6 ESS1 HEF3 HSP104 HXT6 MDN1 PUP2 RPB3 RPN1 RPO21 SPT5 TFG1 TOM1 UTP22 YHR033W |
| View Details | Qiu et al. (2008) |

|
CDC73 CHD1 ELA1 ELP6 IKI1 LEO1 PAF1 RTF1 SPN1 SPT16 SPT4 SPT5 |
The following runs contain data for this protein:
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
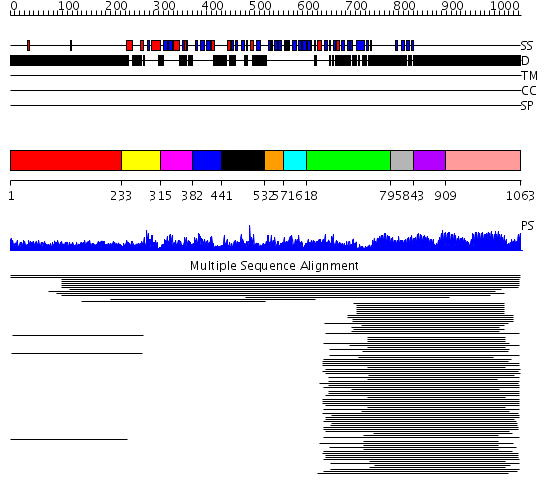
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..232] | 37.191997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [233..314] | 13.366532 | Supt5 repeat No confident structure predictions are available. | |
| 3 | View Details | [315..381] | 15.09691 | Supt5 repeat No confident structure predictions are available. | |
| 4 | View Details | [382..440] | 5.200659 | KOW motif No confident structure predictions are available. | |
| 5 | View Details | [441..531] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [532..570] | 2.744727 | KOW motif No confident structure predictions are available. | |
| 7 | View Details | [571..617] | 4.0 | KOW motif No confident structure predictions are available. | |
| 8 | View Details | [618..794] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [795..842] | 5.0 | KOW motif No confident structure predictions are available. | |
| 10 | View Details | [843..908] | N/A | No confident structure predictions are available. | |
| 11 | View Details | [909..1063] | 3.69897 | Fibrinogen |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.50 |
Source: Reynolds et al. (2008)