













| General Information: |
|
| Name(s) found: |
KIN2 /
YLR096W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1147 amino acids |
Gene Ontology: |
|
| Cellular Component: |
membrane fraction
[IDA plasma membrane [IPI |
| Biological Process: |
exocytosis
[IGI |
| Molecular Function: |
protein kinase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPNPNTADYL VNPNFRTSKG GSLSPTPEAF NDTRVAAPAT LRMMGKQSGP RNDQQQAPLM 60
61 PPADIKQGKE QAAQRQNDAS RPNGAVELRQ FHRRSLGDWE FLETVGAGSM GKVKLVKHRQ 120
121 TKEICVIKIV NRASKAYLHK QHSLPSPKNE SEILERQKRL EKEIARDKRT VREASLGQIL 180
181 YHPHICRLFE MCTMSNHFYM LFEYVSGGQL LDYIIQHGSL KEHHARKFAR GIASALQYLH 240
241 ANNIVHRDLK IENIMISSSG EIKIIDFGLS NIFDYRKQLH TFCGSLYFAA PELLKAQPYT 300
301 GPEVDIWSFG IVLYVLVCGK VPFDDENSSI LHEKIKKGKV DYPSHLSIEV ISLLTRMIVV 360
361 DPLRRATLKN VVEHPWMNRG YDFKAPSYVP NRVPLTPEMI DSQVLKEMYR LEFIDDIEDT 420
421 RRSLIRLVTE KEYIQLSQEY WDKLSNAKGL SSSLNNNYLN STAQQTLIQN HITSNPSQSG 480
481 YNEPDSNFED PTLAYHPLLS IYHLVSEMVA RKLAKLQRRQ ALALQAQAQQ RQQQQQVALG 540
541 TKVALNNNSP DIMTKMRSPQ KEVVPNPGIF QVPAIGTSGT SNNTNTSNKP PLHVMVPPKL 600
601 TIPEQAHTSP TSRKSSDIHT ELNGVLKSTP VPVSGEYQQR SASPVVGEHQ EKNTIGGIFR 660
661 RISQSGQSQH PTRQQEPLPE REPPTYMSKS NEISIKVPKS HSRTISDYIP SARRYPSYVP 720
721 NSVDVKQKPA KNTTIAPPIR SVSQKQNSDL PALPQNAELI VQKQRQKLLQ ENLDKLQIND 780
781 NDNNNVNAVV DGINNDNSDH YLSVPKGRKL HPSARAKSVG HARRESLKFT RPPIPAALPP 840
841 SDMTNDNGFL GEANKERYNP VSSNFSTVPE DSTTYSNDTN NRLTSVYSQE LTEKQILEEA 900
901 SKAPPGSMPS IDYPKSMFLK GFFSVQTTSS KPLPIVRHNI ISVLTRMNID FKEVKGGFIC 960
961 VQQRPSIETA AVPVITTTGV GLDSGKAMDL QNSLDSQLSS SYHSTASSAS RNSSIKRQGS1020
1021 YKRGQNNIPL TPLATNTHQR NSSIPMSPNY GNQSNGTSGE LSSMSLDYVQ QQDDILTTSR1080
1081 AQNINNVNGQ TEQTNTSGIK ERPPIKFEIH IVKVRIVGLA GVHFKKVSGN TWLYKELASY1140
1141 ILKELNL |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
AIM3 BAR1 COA4 GYL1 GYP5 HNT1 KIN2 MEF2 NCP1 PCL10 PRP28 RVS161 RVS167 SAE2 SLK19 TIM21 VPS15 |
| View Details | Ho Y, et al. (2002) |

|
AHA1 ARP2 BCK1 BUD14 CMP2 CPR6 DOG1 EGD2 FOL2 GAL7 GIS4 GND1 HSP104 IDH1 ILV5 IPP1 KEL1 KEL2 KIC1 KIN2 KRE6 LHS1 LYS12 MKK2 MKT1 OYE2 PDC6 PIL1 PMA1 POP2 QCR2 RCN2 RPN6 RPT3 SIS1 SKG3 SLT2 SMK1 TEF4 TFC4 TIF1, TIF2 UBA1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample bir 1 from feb 2005 | Sandall S, et all (2006) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
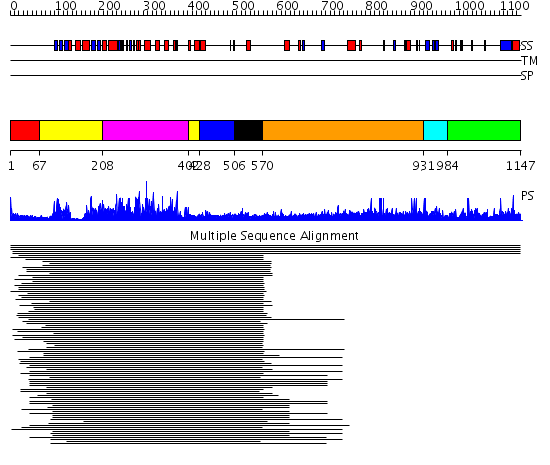
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..66] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [67..207] [402..427] |
458.08661 | Calmodulin-dependent protein kinase | |
| 3 | View Details | [208..401] | 458.08661 | Calmodulin-dependent protein kinase | |
| 4 | View Details | [428..505] | 19.522879 | Calmodulin | |
| 5 | View Details | [506..569] | 19.522879 | Calmodulin | |
| 6 | View Details | [570..930] | 9.14 | RBP1 | |
| 7 | View Details | [931..983] | 8.1 | Sec24 | |
| 8 | View Details | [984..1147] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)