













| Protein: | KIN2 |
| From Publication: | Ho Y. et al. (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002 Jan 10;415(6868):180-3. |
| Complexes containing KIN2: | 2 |
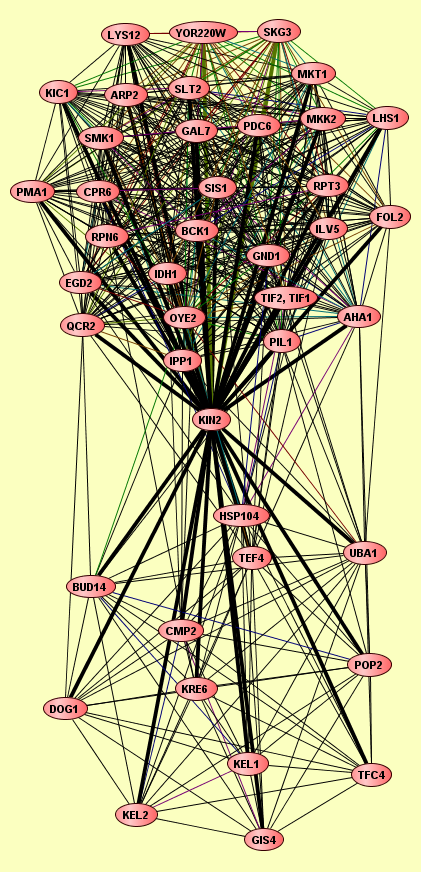
| LEGEND: |
|||
| = Same process, or one is unknown. | = Same branch, distance 4. | ||
| = Same branch, distance 1. | = Same branch, distance 5. | ||
| = Same branch, distance 2. | = Same branch, distance 6 or more. | ||
| = Same branch, distance 3. | = Not in same branch of GO. | ||
| Size | Notes | Members | |
| View GO Analysis | 30 proteins | This molecular complex record represents a population of complexes co-purified by immunoprecipitation of FLAG-tagged Slt2. The topolgies of protein complexes in this experiment are unknown. | AHA1 ARP2 BCK1 CPR6 EGD2 FOL2 GAL7 GND1 IDH1 ILV5 IPP1 KIC1 KIN2 LHS1 LYS12 MKK2 MKT1 OYE2 PDC6 PIL1 PMA1 QCR2 RCN2 RPN6 RPT3 SIS1 SKG3 SLT2 SMK1 TIF1, TIF2 |
| View GO Analysis | 13 proteins | This molecular complex record represents a population of complexes co-purified by immunoprecipitation of FLAG-tagged Kin2. The topolgies of protein complexes in this experiment are unknown. | BUD14 CMP2 DOG1 GIS4 HSP104 KEL1 KEL2 KIN2 KRE6 POP2 TEF4 TFC4 UBA1 |