













| General Information: |
|
| Name(s) found: |
ABF1 /
YKL112W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 731 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA nuclear chromatin [TAS nucleotide-excision repair factor 4 complex [IPI |
| Biological Process: |
negative regulation of transcription from RNA polymerase II promoter
[IMP chromatin remodeling [IMP nucleotide-excision repair, DNA damage recognition [IDA DNA replication [IDA chromatin silencing at silent mating-type cassette [IMP positive regulation of transcription from RNA polymerase II promoter [IMP |
| Molecular Function: |
DNA binding
[IDA transcription factor activity [TAS DNA bending activity [IDA chromatin binding [IDA transcription activator activity [IDA transcription repressor activity [IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDKLVVNYYE YKHPIINKDL AIGAHGGKKF PTLGAWYDVI NEYEFQTRCP IILKNSHRNK 60
61 HFTFACHLKN CPFKVLLSYA GNAASSETSS PSANNNTNPP GTPDHIHHHS NNMNNEDNDN 120
121 NNGSNNKVSN DSKLDFVTDD LEYHLANTHP DDTNDKVESR SNEVNGNNDD DADANNIFKQ 180
181 QGVTIKNDTE DDSINKASID RGLDDESGPT HGNDSGNHRH NEEDDVHTQM TKNYSDVVND 240
241 EDINVAIANA VANVDSQSNN KHDGKDDDAT NNNDGQDNNT NNDHNNNSNI NNNNVGSHGI 300
301 SSHSPSSIRD TSMNLDVFNS ATDDIPGPFV VTKIEPYHSH PLEDNLSLGK FILTKIPKIL 360
361 QNDLKFDQIL ESSYNNSNHT VSKFKVSHYV EESGLLDILM QRYGLTAEDF EKRLLSQIAR 420
421 RITTYKARFV LKKKKMGEYN DLQPSSSSNN NNNNDGELSG TNLRSNSIDY AKHQEISSAG 480
481 TSSNTTKNVN NNKNDSNDDN NGNNNNDASN LMESVLDKTS SHRYQPKKMP SVNKWSKPDQ 540
541 ITHSDVSMVG LDESNDGGNE NVHPTLAEVD AQEARETAQL AIDKINSYKR SIDDKNGDGH 600
601 NNSSRNVVDE NLINDMDSED AHKSKRQHLS DITLEERNED DKLPHEVAEQ LRLLSSHLKE 660
661 VENLHQNNDD DVDDVMVDVD VESQYNKNTT HHNNHHSQPH HDEEDVAGLI GKADDEEDLS 720
721 DENIQPELRG Q |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
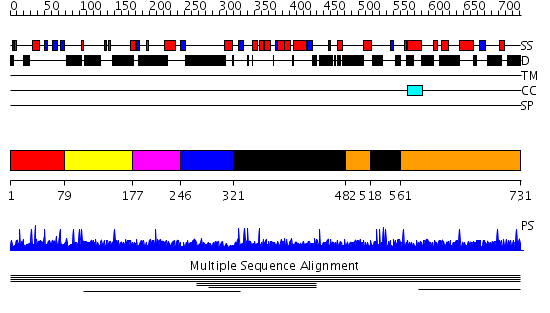
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..78] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [79..176] | 7.56 | No description for 1l8mA was found. | |
| 3 | View Details | [177..245] | 7.56 | No description for 1l8mA was found. | |
| 4 | View Details | [246..320] | 7.56 | No description for 1l8mA was found. | |
| 5 | View Details | [321..481] [518..560] |
7.09 | Monomeric isocitrate dehydrogenase | |
| 6 | View Details | [482..517] [561..731] |
7.09 | Monomeric isocitrate dehydrogenase |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)