













| General Information: |
|
| Name(s) found: |
BCK1 /
YJL095W
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1478 amino acids |
Gene Ontology: |
|
| Cellular Component: |
intracellular
[TAS |
| Biological Process: |
response to acid
[IMP endoplasmic reticulum unfolded protein response [IMP intracellular protein kinase cascade [IMP response to nutrient [IMP protein amino acid phosphorylation [TAS |
| Molecular Function: |
MAP kinase kinase kinase activity
[IGI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPFLRKIAGT AHTHSRSDSN SSVKFGHQPT SSVASTKSSS KSPRATSRKS IYDDIRSQFP 60
61 NLTPNSTSSQ FYESTPVIEQ SFNWTTDDHI SAGTLENPTS FTNSSYKNDN GPSSLSDSRK 120
121 SSGGNSVNSL SFDKLILSWD PTDPDEWTMH RVTSWFKFHD FPESWILFFK KHQLFGHRFI 180
181 KLLAYDNFAV YEKYLPQTKT ASYTRFQQLL KKTMTKNVTN SHIRQKSASK LKSSRSSSES 240
241 IKSKLKNSKS QEDISNSRST SESALSPTKS GPSKTDEKNF LHSTSTHQKT KSASSLYRRS 300
301 FISLRGSSSS NASSAKSPSN IKLSIPARPH SIIESNSTLT KSASPPASPS YPSIFRRHHK 360
361 SSSSESSLLN SLFGSGIGEE APTKPNPQGH SLSSENLAKG KSKHYETNVS SPLKQSSLPT 420
421 SDDKGNLWNK FKRKSQIGVP SPNTVAYVTS QETPSLKSNS STATLTVQTA DVNIPSPSSS 480
481 PPPIPKTANR SLEVISTEDT PKISSTTASF KETYPDCINP DKTVPVPVNN QKYSVKNFLL 540
541 DQKFYPLKKT GLNDSENKYI LVTKDNVSFV PLNLKSVAKL SSFKESALTK LGINHKNVTF 600
601 HMTDFDCDIG AAIPDDTLEF LKKSLFLNTS GKIYIKDQMK LQQKPKPAPL TSENNVPLKS 660
661 VKSKSSMRSG TSSLIASTDD VSIVTSSSDI TSFDEHASGS GRRYPQTPSY YYDRVSNTNP 720
721 TEELNYWNIK EVLSHEENAP KMVFKTSPKL ELNLPDKGSK LNIPTPITEN ESKSSFQVLR 780
781 KDEGTEIDFN HRRESPYTKP ELAPKREAPK PPANTSPQRT LSTSKQNKPI RLVRASTKIS 840
841 RSKRSKPLPP QLLSSPIEAS SSSPDSLTSS YTPASTHVLI PQPYKGANDV MRRLKTDQDS 900
901 TSTSPSLKMK QKVNRSNSTV STSNSIFYSP SPLLKRGNSK RVVSSTSAAD IFEENDITFA 960
961 DAPPMFDSDD SDDDSSSSDD IIWSKKKTAP ETNNENKKDE KSDNSSTHSD EIFYDSQTQD1020
1021 KMERKMTFRP SPEVVYQNLE KFFPRANLDK PITEGIASPT SPKSLDSLLS PKNVASSRTE1080
1081 PSTPSRPVPP DSSYEFIQDG LNGKNKPLNQ AKTPKRTKTI RTIAHEASLA RKNSVKLKRQ1140
1141 NTKMWGTRMV EVTENHMVSI NKAKNSKGEY KEFAWMKGEM IGKGSFGAVY LCLNVTTGEM1200
1201 MAVKQVEVPK YSSQNEAILS TVEALRSEVS TLKDLDHLNI VQYLGFENKN NIYSLFLEYV1260
1261 AGGSVGSLIR MYGRFDEPLI KHLTTQVLKG LAYLHSKGIL HRDMKADNLL LDQDGICKIS1320
1321 DFGISRKSKD IYSNSDMTMR GTVFWMAPEM VDTKQGYSAK VDIWSLGCIV LEMFAGKRPW1380
1381 SNLEVVAAMF KIGKSKSAPP IPEDTLPLIS QIGRNFLDAC FEINPEKRPT ANELLSHPFS1440
1441 EVNETFNFKS TRLAKFIKSN DKLNSSKLRI TSQENKTE |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
BCK1 YIP5 |
| View Details | Gavin AC, et al. (2002) |
|
ACE2 ALT2 BCK1 CDC11 DRE2 ECM30 ENP2 IML1 MNL1 MTR4 MYO2 RGC1 SEC3 UBP15 YBL044W YJR098C YNL313C |
| View Details | Ho Y, et al. (2002) |
|
AHA1 ARO1 ARP2 BCK1 BUL1 CHS1 CMP2 CPR6 DBP7 EGD2 FOL2 GAL7 GND1 IDH1 ILV5 IPP1 KIC1 KIN2 KOG1 KSP1 LHS1 LYS12 MKK2 MKT1 NPA3 OYE2 PDC6 PIL1 PMA1 PRB1 PRI2 PSY2 QCR2 RCN2 RGD1 RNQ1 RPN1 RPN6 RPN7 RPT3 RVB1 SIS1 SKG3 SLT2 SMK1 TIF1, TIF2 TPD3 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
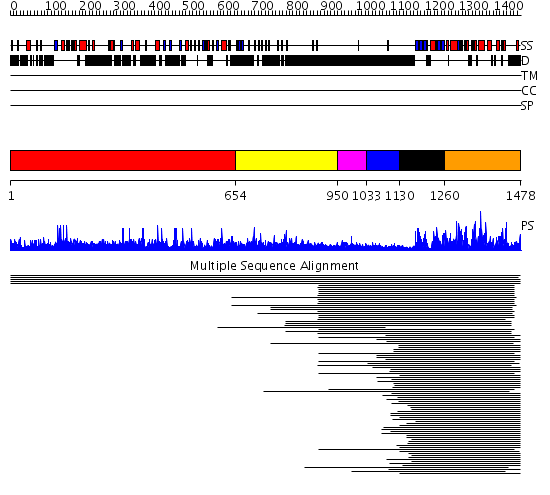
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..653] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [654..949] | 10.04 | No description for 1ldvA was found. | |
| 3 | View Details | [950..1032] | 15.0 | pak1 autoregulatory domain | |
| 4 | View Details | [1033..1129] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [1130..1259] | 486.9897 | pak1 | |
| 6 | View Details | [1260..1478] | 486.9897 | pak1 | |
| 7 | View Details | [808..921] | 10.30103 | No description for 2nptB was found. | |
| 8 | View Details | [922..1087] | 16.221849 | pak1 autoregulatory domain | |
| 9 | View Details | [1088..1171] | 34.30103 | No description for 2ozoA was found. | |
| 10 | View Details | [1172..1478] | 117.0 | Catalytic and ubiqutin-associated domains of MARK2/PAR-1: Wild type |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 10 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)