













| General Information: |
|
| Name(s) found: |
PYC2 /
YBR218C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1180 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[IDA |
| Biological Process: |
gluconeogenesis
[IMP NADPH regeneration [TAS] |
| Molecular Function: |
pyruvate carboxylase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSSKKLAGL RDNFSLLGEK NKILVANRGE IPIRIFRSAH ELSMRTIAIY SHEDRLSMHR 60
61 LKADEAYVIG EEGQYTPVGA YLAMDEIIEI AKKHKVDFIH PGYGFLSENS EFADKVVKAG 120
121 ITWIGPPAEV IDSVGDKVSA RHLAARANVP TVPGTPGPIE TVQEALDFVN EYGYPVIIKA 180
181 AFGGGGRGMR VVREGDDVAD AFQRATSEAR TAFGNGTCFV ERFLDKPKHI EVQLLADNHG 240
241 NVVHLFERDC SVQRRHQKVV EVAPAKTLPR EVRDAILTDA VKLAKVCGYR NAGTAEFLVD 300
301 NQNRHYFIEI NPRIQVEHTI TEEITGIDIV SAQIQIAAGA TLTQLGLLQD KITTRGFSIQ 360
361 CRITTEDPSK NFQPDTGRLE VYRSAGGNGV RLDGGNAYAG ATISPHYDSM LVKCSCSGST 420
421 YEIVRRKMIR ALIEFRIRGV KTNIPFLLTL LTNPVFIEGT YWTTFIDDTP QLFQMVSSQN 480
481 RAQKLLHYLA DLAVNGSSIK GQIGLPKLKS NPSVPHLHDA QGNVINVTKS APPSGWRQVL 540
541 LEKGPSEFAK QVRQFNGTLL MDTTWRDAHQ SLLATRVRTH DLATIAPTTA HALAGAFALE 600
601 CWGGATFDVA MRFLHEDPWE RLRKLRSLVP NIPFQMLLRG ANGVAYSSLP DNAIDHFVKQ 660
661 AKDNGVDIFR VFDALNDLEQ LKVGVNAVKK AGGVVEATVC YSGDMLQPGK KYNLDYYLEV 720
721 VEKIVQMGTH ILGIKDMAGT MKPAAAKLLI GSLRTRYPDL PIHVHSHDSA GTAVASMTAC 780
781 ALAGADVVDV AINSMSGLTS QPSINALLAS LEGNIDTGIN VEHVRELDAY WAEMRLLYSC 840
841 FEADLKGPDP EVYQHEIPGG QLTNLLFQAQ QLGLGEQWAE TKRAYREANY LLGDIVKVTP 900
901 TSKVVGDLAQ FMVSNKLTSD DIRRLANSLD FPDSVMDFFE GLIGQPYGGF PEPLRSDVLR 960
961 NKRRKLTCRP GLELEPFDLE KIREDLQNRF GDIDECDVAS YNMYPRVYED FQKIRETYGD1020
1021 LSVLPTKNFL APAEPDEEIE VTIEQGKTLI IKLQAVGDLN KKTGQREVYF ELNGELRKIR1080
1081 VADKSQNIQS VAKPKADVHD THQIGAPMAG VIIEVKVHKG SLVKKGESIA VLSAMKMEMV1140
1141 VSSPADGQVK DVFIKDGESV DASDLLVVLE EETLPPSQKK |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
PYC1 PYC2 |
| View Details | Krogan NJ, et al. (2006) |
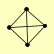
|
PYC1 PYC2 SPS1 TPM1 |
| View Details | Ho Y, et al. (2002) |
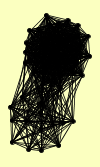
|
AAC3 ACC1 ACH1 ADH2 ADR1 AHP1 CAR2 FOL2 GCD11 GFA1 GND1 GUS1 HYP2 IDH2 ILV5 ISA2 MDH1 MET18 MPC54 NGL2 OYE2 OYE3 POR1 PST2 PYC1 PYC2 RET1 RPA135 RPA190 RPA49 RPB5 RPC19 RPC25 RPC34 RPC40 RPC82 RPN3 RPO26 RPO31 RVS167 SEC27 SOD1 TBS1 TFP1 TSA2 YBL108W YCK1 YFL042C YHR112C |
| View Details | Qiu et al. (2008) |
|
PYC1 PYC2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi 1 with ha tag from october 2005 | McCusker D, et al (2007) | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
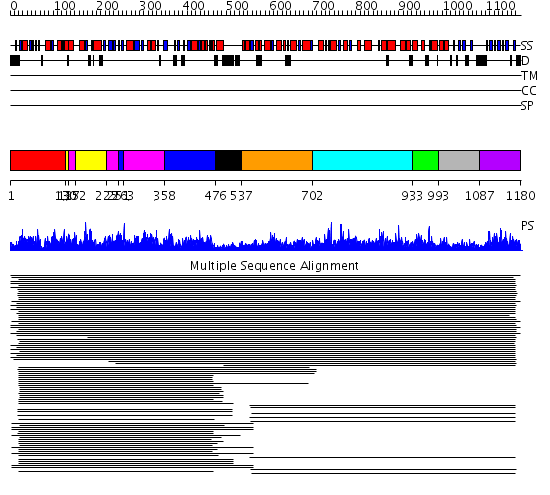
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..129] | 1114.228787 | No description for 1lsrA_ was found. | |
| 2 | View Details | [130..136] [152..224] |
1114.228787 | No description for 1lsrA_ was found. | |
| 3 | View Details | [137..151] [225..250] [263..357] |
1114.228787 | No description for 1lsrA_ was found. | |
| 4 | View Details | [251..262] [358..475] |
1114.228787 | No description for 1lsrA_ was found. | |
| 5 | View Details | [476..536] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [537..701] | 9.05 | Betaine-homocysteine S-methyltransferase | |
| 7 | View Details | [702..932] | 9.79 | Dihydrodipicolinate synthase | |
| 8 | View Details | [933..992] | 9.79 | Dihydrodipicolinate synthase | |
| 9 | View Details | [993..1086] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1087..1180] | 86.39794 | Biotin carboxyl carrier domain of transcarboxylase (TC 1.3S) |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)