













| General Information: |
|
| Name(s) found: |
IPI3 /
YNL182C
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 555 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA nuclear outer membrane [IMP nucleoplasm [IDA |
| Biological Process: |
ribosome biogenesis
[RCA ribosomal large subunit assembly [IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDEQVIFTTN TSGTIASVHS FEQINLRQCS TQSRNSCVQV GNKYLFIAQA QKALINVYNL 60
61 SGSFKRESVE QRLPLPEILK CLEVVENDGV QYDRIQGVNH NLPDFNLPYL LLGSTESGKL 120
121 YIWELNSGIL LNVKPMAHYQ SITKIKSILN GKYIITSGND SRVIIWQTVD LVSASNDDPK 180
181 PLCILHDHTL PVTDFQVSSS QGKFLSCTDT KLFTVSQDAT IRCYDLSLIG SKKKQKANEN 240
241 DVSIGKTPVL LATFTTPYSI KSIVLDPADR ACYIGTAEGC FSLNLFYKLK GNAIVNLLQS 300
301 AGVNTVQKGR VFSLVQRNSL TGGENEDLDA LYAMGQLVCE NVLNSNVSCL EISMDGTLLL 360
361 IGDTEGKVSI AEIYSKQIIR TIQTLTTSQD SVGEVTNLLT NPYRLERGNL LFEGESKGKQ 420
421 PSNNNGHNFM KIPNLQRVIF DGKNKGHLHD IWYQIGEPEA ETDPNLALPL NDFNAYLEQV 480
481 KTQESIFSHI GKVSSNVKVI DNKIDATSSL DSNAAKDEEI TELKTNIEAL THAYKELRDM 540
541 HEKLYEEHQQ MLDKQ |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
ARX1 BUD20 CIC1 DBP10 IPI1 IPI3 MDN1 MRT4 NOG1 NOG2 NOP15 NOP7 NSA2 NUG1 RIX1 RLP7 RPF1 RSA4 SDA1 TIF6 |
| View Details | Hazbun TR, et al. (2003) |
|
IPI1 IPI3 RIX1 |
| View Details | Krogan NJ, et al. (2006) |

|
IPI1 IPI3 RIX1 |
| View Details | Gavin AC, et al. (2006) |

|
BUD20 IPI3 NOG1 NOG2 NSA2 NUG1 RIX1 RPF2 RSA4 SDA1 |
| View Details | Gavin AC, et al. (2002) |

|
ARX1 BEM2 BMS1 BRX1 BUD20 CIC1 CLU1 CYR1 CYS4 DBP10 DBP9 DCP2 DRS1 EBP2 ERB1 FPR4 GCN1 HAS1 HDA1 HER1 HIS3 INP2 IPI1 IPI3 KRE33 LHP1 LOC1 LSG1 MAK21 MAK5 MDN1 MIS1 MRT4 NAP1 NIP1 NIP7 NMD3 NOC2 NOC3 NOG1 NOG2 NOP1 NOP12 NOP15 NOP16 NOP2 NOP4 NOP56 NOP7 NSA1 NSA2 NUG1 PRT1 PUF6 REI1 RIX1 RIX7 RLP24 RLP7 RPA135 RPA190 RPF1 RPF2 RPG1 RPP2B RPT6 RRP1 RRP12 RSA3 RSA4 RTR1 SDA1 SEC7 SKI2 SNU13 SPB1 SPB4 SSF1 TIF6 TUB3 URB1 UTP10 UTP20 UTP22 YGL036W YTM1 |
| View Details | Ho Y, et al. (2002) |
|
ATG17 ATP3 BUD3 HHF1, HHF2 HTB2 IPI3 Q0032 RIX1 RPC19 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #20 Asynchronous Prep1-TiO2 Phosphopeptide enrichment, PartB | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Hazbun TR, et al. (2003) | ||
| View Screen | 2 | Hazbun TR, et al. (2003) | ||
| View Screen | 2 | Hazbun TR, et al. (2003) |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Hazbun TR, et al. (2003) |
| View Data |
|
Unpublished Data |
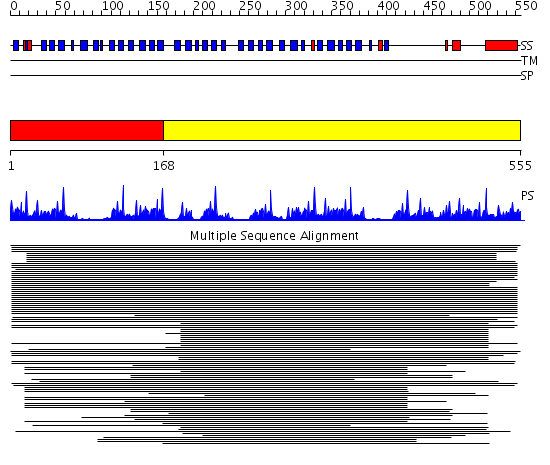
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..167] | 56.522879 | Tup1, C-terminal domain | |
| 2 | View Details | [168..555] | 52.221849 | beta1-subunit of the signal-transducing G protein heterotrimer |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)