













| General Information: |
|
| Name(s) found: |
IMD2 /
YHR216W
[SGD]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 523 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
GTP biosynthetic process
[TAS |
| Molecular Function: |
IMP dehydrogenase activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAAIRDYKTA LDFTKSLPRP DGLSVQELMD SKIRGGLTYN DFLILPGLVD FASSEVSLQT 60
61 KLTRNITLNI PLVSSPMDTV TESEMATFMA LLGGIGFIHH NCTPEDQADM VRRVKNYENG 120
121 FINNPIVISP TTTVGEAKSM KEKYGFAGFP VTTDGKRNAK LVGVITSRDI QFVEDNSLLV 180
181 QDVMTKNPVT GAQGITLSEG NEILKKIKKG RLLVVDEKGN LVSMLSRTDL MKNQNYPLAS 240
241 KSANTKQLLC GASIGTMDAD KERLRLLVKA GLDVVILDSS QGNSIFELNM LKWVKESFPG 300
301 LEVIAGNVVT REQAANLIAA GADGLRIGMG TGSICITQEV MACGRPQGTA VYNVCEFANQ 360
361 FGVPCMADGG VQNIGHITKA LALGSSTVMM GGMLAGTTES PGEYFYQDGK RLKAYRGMGS 420
421 IDAMQKTGTK GNASTSRYFS ESDSVLVAQG VSGAVVDKGS IKKFIPYLYN GLQHSCQDIG 480
481 CRSLTLLKNN VQRGKVRFEF RTASAQLEGG VHNLHSYEKR LHN |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
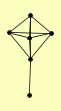
|
IMD1 IMD2 IMD3 IMD4 RPL13B YAR075W |
| View Details | Krogan NJ, et al. (2006) |
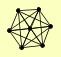
|
COX18 IMD1 IMD2 IMD3 IMD4 PDE2 YAR075W |
| View Details | Gavin AC, et al. (2006) |
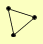
|
IMD2 LYS2 SXM1 |
| View Details | Ho Y, et al. (2002) |

|
AAC3 AI1 AIM46 ARE1 BUD32 CAR2 CDC33 CIC1 CKA1 CKA2 CKB1 CPR6 CRM1 DBP7 DIA4 DRS1 EBP2 FET4 FUN12 GRS1 GRX3 GRX4 HAS1 HEF3 HHO1 IDP2 IMD2 IMD3 IMD4 KAE1 KRE33 KRI1 MAG1 MAG2 MGM101 MPH1 MSC3 MSH2 NOC2 NOP1 NOP12 NOP13 NOP4 NOP7 NPA3 NPL3 OYE2 PDC6 PHO81 PUF6 RET1 RNH202 RPC82 RPF2 RPN1 RPN5 RPN6 RPT1 RRP12 RRP5 SAH1 SEC18 SEC23 SGO1 SPT2 TIF4631 TIF4632 TMA29 URA7 UTP22 YHR033W |
The following runs contain data for this protein:
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
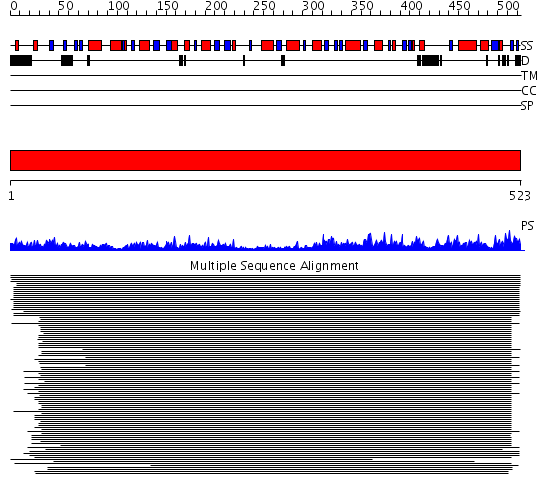
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..523] | 1947.0 | Inosine monophosphate dehydrogenase (IMPDH) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)