













| General Information: |
|
| Name(s) found: |
HSP60 /
YLR259C
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 572 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA mitochondrial nucleoid [IDA |
| Biological Process: |
protein import into mitochondrial matrix
[TAS protein folding [TAS |
| Molecular Function: |
single-stranded DNA binding
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLRSSVVRSR ATLRPLLRRA YSSHKELKFG VEGRASLLKG VETLAEAVAA TLGPKGRNVL 60
61 IEQPFGPPKI TKDGVTVAKS IVLKDKFENM GAKLLQEVAS KTNEAAGDGT TSATVLGRAI 120
121 FTESVKNVAA GCNPMDLRRG SQVAVEKVIE FLSANKKEIT TSEEIAQVAT ISANGDSHVG 180
181 KLLASAMEKV GKEGVITIRE GRTLEDELEV TEGMRFDRGF ISPYFITDPK SSKVEFEKPL 240
241 LLLSEKKISS IQDILPALEI SNQSRRPLLI IAEDVDGEAL AACILNKLRG QVKVCAVKAP 300
301 GFGDNRKNTI GDIAVLTGGT VFTEELDLKP EQCTIENLGS CDSITVTKED TVILNGSGPK 360
361 EAIQERIEQI KGSIDITTTN SYEKEKLQER LAKLSGGVAV IRVGGASEVE VGEKKDRYDD 420
421 ALNATRAAVE EGILPGGGTA LVKASRVLDE VVVDNFDQKL GVDIIRKAIT RPAKQIIENA 480
481 GEEGSVIIGK LIDEYGDDFA KGYDASKSEY TDMLATGIID PFKVVRSGLV DASGVASLLA 540
541 TTEVAIVDAP EPPAAAGAGG MPGGMPGMPG MM |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
ASE1 BFR1 HSP60 |
| View Details | Hazbun TR, et al. (2003) |

|
HSP60 TAM41 |
| View Details | Krogan NJ, et al. (2006) |
|
ALT1 HSP60 YPL247C |
| View Details | Qiu et al. (2008) |
|
HSP60 HSP78 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Cheeseman IM, et al. (2002) | |
| View Run | No Comments | Cheeseman IM, et al. (2002) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Cheeseman IM, et al. (2002) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | De Craene, J., et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | No Comments | Green EM, et al (2005) | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | experimental1 - mih1-1 | McCusker D, et al (2007) | |
| View Run | experimental2 - mih1-2 | McCusker D, et al (2007) | |
| View Run | 3908: TAP-tagged, includes hir2(delta) in background | Green EM, et al (2005) | |
| View Run | 3912: TAP-tagged, strain background includes hpc2(delta) | Green EM, et al (2005) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample zds1 from august 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi 1 with ha tag from october 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi1 with gst from october 2005 | McCusker D, et al (2007) | |
| View Run | Sample bob1 (2nd set) from october 2005 | McCusker D, et al (2007) | |
| View Run | Boi 2 gst control | McCusker D, et al (2007) | |
| View Run | Sample: HX11 - both fusion and trans-SNARE complex formation take place. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | Sample: HX14 - mixture in detergent (control). | Hao Xu, et al. (2010) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #17 Mitotic Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
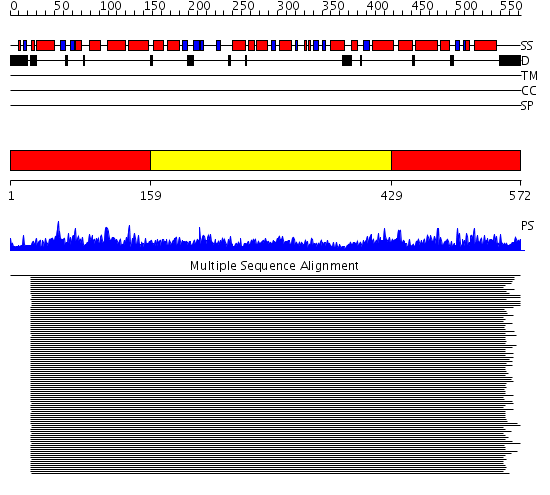
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..158] [429..572] |
2510.0 | GroEL, E domain; GroEL, A domain; GroEL, I domain | |
| 2 | View Details | [159..428] | 2510.0 | GroEL, E domain; GroEL, A domain; GroEL, I domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)