













| General Information: |
|
| Name(s) found: |
NUP100 /
YKL068W
[SGD]
|
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 959 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear pore
[IDA |
| Biological Process: |
mRNA export from nucleus
[TAS rRNA export from nucleus [TAS nuclear pore organization [TAS snRNP protein import into nucleus [TAS mRNA-binding (hnRNP) protein import into nucleus [TAS snRNA export from nucleus [TAS tRNA export from nucleus [TAS NLS-bearing substrate import into nucleus [TAS protein export from nucleus [TAS ribosomal protein import into nucleus [TAS |
| Molecular Function: |
structural molecule activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFGNNRPMFG GSNLSFGSNT SSFGGQQSQQ PNSLFGNSNN NNNSTSNNAQ SGFGGFTSAA 60
61 GSNSNSLFGN NNTQNNGAFG QSMGATQNSP FGSLNSSNAS NGNTFGGSSS MGSFGGNTNN 120
121 AFNNNSNSTN SPFGFNKPNT GGTLFGSQNN NSAGTSSLFG GQSTSTTGTF GNTGSSFGTG 180
181 LNGNGSNIFG AGNNSQSNTT GSLFGNQQSS AFGTNNQQGS LFGQQSQNTN NAFGNQNQLG 240
241 GSSFGSKPVG SGSLFGQSNN TLGNTTNNRN GLFGQMNSSN QGSSNSGLFG QNSMNSSTQG 300
301 VFGQNNNQMQ INGNNNNSLF GKANTFSNSA SGGLFGQNNQ QQGSGLFGQN SQTSGSSGLF 360
361 GQNNQKQPNT FTQSNTGIGL FGQNNNQQQQ STGLFGAKPA GTTGSLFGGN SSTQPNSLFG 420
421 TTNVPTSNTQ SQQGNSLFGA TKLTNMPFGG NPTANQSGSG NSLFGTKPAS TTGSLFGNNT 480
481 ASTTVPSTNG LFGNNANNST STTNTGLFGA KPDSQSKPAL GGGLFGNSNS NSSTIGQNKP 540
541 VFGGTTQNTG LFGATGTNSS AVGSTGKLFG QNNNTLNVGT QNVPPVNNTT QNALLGTTAV 600
601 PSLQQAPVTN EQLFSKISIP NSITNPVKAT TSKVNADMKR NSSLTSAYRL APKPLFAPSS 660
661 NGDAKFQKWG KTLERSDRGS STSNSITDPE SSYLNSNDLL FDPDRRYLKH LVIKNNKNLN 720
721 VINHNDDEAS KVKLVTFTTE SASKDDQASS SIAASKLTEK AHSPQTDLKD DHDESTPDPQ 780
781 SKSPNGSTSI PMIENEKISS KVPGLLSNDV TFFKNNYYIS PSIETLGNKS LIELRKINNL 840
841 VIGHRNYGKV EFLEPVDLLN TPLDTLCGDL VTFGPKSCSI YENCSIKPEK GEGINVRCRV 900
901 TLYSCFPIDK ETRKPIKNIT HPLLKRSIAK LKENPVYKFE SYDPVTGTYS YTIDHPVLT |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | (MIPS) Mewes HW, et al. (2004) |
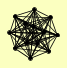
|
GLE2 NOP14 NUP1 NUP100 NUP116 NUP133 NUP157 NUP159 NUP170 NUP188 NUP2 NUP42 POM152 |
| View Details | Qiu et al. (2008) |
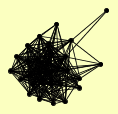
|
GLE1 GLE2 KAP123 MEX67 NDC1 NIC96 NOP14 NSP1 NUP1 NUP100 NUP116 NUP120 NUP133 NUP145 NUP157 NUP159 NUP170 NUP188 NUP192 NUP2 NUP42 NUP49 NUP53 NUP57 NUP82 NUP84 NUP85 POM152 SEC13 SUS1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
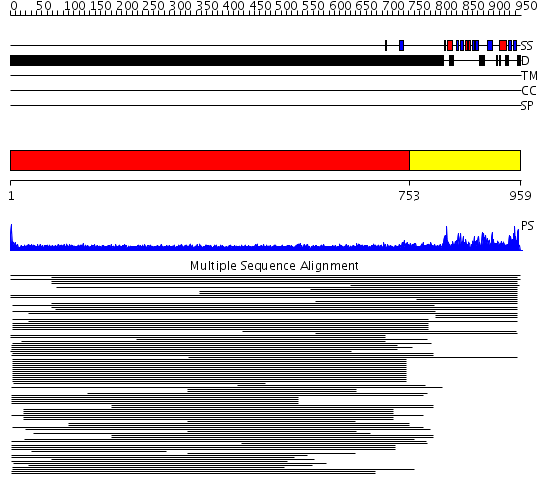
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..752] | 20.175996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [753..959] | 1.105938 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.86 |
Source: Reynolds et al. (2008)