













| General Information: |
|
| Name(s) found: |
DEP1 /
YAL013W
[SGD]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 420 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IC |
| Biological Process: |
telomere maintenance
[IMP phospholipid metabolic process [IMP histone deacetylation [IDA regulation of transcription from RNA polymerase II promoter [IMP |
| Molecular Function: |
histone deacetylase activity
[IDA transcription regulator activity [IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSQQTPQESE QTTAKEQDLD QESVLSNIDF NTDLNHNLNL SEYCISSDAG TEKMDSDEEK 60
61 SLANLPELKY APKLSSLVKQ ETLTESLKRP HEDEKEAIDE AKKMKVPGEN EDESKEEEKS 120
121 QELEEAIDSK EKSTDARDEQ GDEGDNEEEN NEEDNENENE HTAPPALVMP SPIEMEEQRM 180
181 TALKEITDIE YKFAQLRQKL YDNQLVRLQT ELQMCLEGSH PELQVYYSKI AAIRDYKLHR 240
241 AYQRQKYELS CINTETIATR TFIHQDFHKK VTDLRARLLN RTTQTWYDIN KERRDMDIVI 300
301 PDVNYHVPIK LDNKTLSCIT GYASAAQLCY PGEPVAEDLA CESIEYRYRA NPVDKLEVIV 360
361 DRMRLNNEIS DLEGLRKYFH SFPGAPELNP LRDSEINDDF HQWAQCDRHT GPHTTSFCYS 420
421 |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
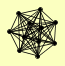
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
CTI6 DEP1 EAF3 PHO23 RCO1 RPD3 RXT2 SAP30 SDS3 SIN3 UME1 |
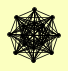
| View Details | Krogan NJ, et al. (2006) |
|
ASH1 COQ6 CTI6 DEP1 EAF3 GRX5 PHO23 RCO1 RPD3 RXT2 RXT3 SAP30 SAY1 SDS3 SIN3 UME1 |
| View Details | Gavin AC, et al. (2002) |

|
ABF2 CKA1 CKA2 CKB1 CKB2 CTI6 DEP1 DOT6 EAF3 ECM16 HTA2 IOC3 ISW1 MED4 NHP10 NRD1 PHO23 RCO1 RPD3 RSC8 RXT2 RXT3 SAP30 SDS3 SIN3 STO1 TOP1 TUP1 UME1 UME6 VPS1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data

Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..183] | 1.087995 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [184..420] | N/A | Confident ab initio structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)