













| General Information: |
|
| Name(s) found: |
BEM3 /
YPL115C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1128 amino acids |
Gene Ontology: |
|
| Cellular Component: |
intracellular
[TAS |
| Biological Process: |
budding cell isotropic bud growth
[IPI small GTPase mediated signal transduction [IPI pseudohyphal growth [IPI invasive growth in response to glucose limitation [IPI budding cell apical bud growth [IPI |
| Molecular Function: |
Rho GTPase activator activity
[IDA phosphatidylinositol-3-phosphate binding [IDA signal transducer activity [IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTDNLTTTHG GSTTLELLAQ YNDHRSKKDK SIEHIEKGTC SGKERNPSYD EIFTENIKLK 60
61 LQVQEYETEI ESLEKVIDML QKNREASLEV VLEQVQNDSR DSYVNDQSFV LPPRSAERKA 120
121 HIKSLNLPIP TLSPPLQQGS DVALETSVTP TVPQIGVTSN TSISRKHLQN MILNDEIEAN 180
181 SSFSSPKIIN RSVSSPTKIH SEQLASPAAS VTYTTSRITI KSPNKGSKSP LQERLRSPQN 240
241 PNRMTAVINN HLHSPLKAST SNNLDELTES KSQQLTNDAI QKNDRVYSSI TSSAYTTGTP 300
301 TSAAKSPSSL LEVKEGENKA LGFSPASKEK LDDFTQLLDS SFGEEDLVNT DSKDPLSIKS 360
361 TINESLPPPP APPTFFSPTS SGNIKNSTPL SSHLASPVIL NKKDDNFGAQ SAKNLKKPVL 420
421 TSSLPNLSTK LSTTSQNASL PPNPPVESSS KQKQLGETAS IHSTNTLNTF SSTPQGSLKT 480
481 LRRPHASSVS TVKSVAQSLK SDIPLFVQPE DFGTIQIEVL STLYRDNEDD LSILIAIIDR 540
541 KSGKEMFKFS KSIHKVRELD VYMKSHVPDL PLPTLPDRQL FQTLSPTKVD TRKNILNQYY 600
601 TSIFSVPEFP KNVGLKIAQF ISTDTVMTPP MMDDNVKDGS LLLRRPKTLT GNSTWRVRYG 660
661 ILRDDVLQLF DKNQLTETIK LRQSSIELIP NLPEDRFGTR NGFLITEHKK SGLSTSTKYY 720
721 ICTETSKERE LWLSAFSDYI DPSQSLSLSS SRNANDTDSA SHLSAGTHHS KFGNATISAT 780
781 DTPSYVTDLT QEYNNNNNIS NSSNNIANSD GIDSNPSSHS NFLASSSGNA EEEKDSRRAK 840
841 MRSLFPFKKL TGPASAMNHI GITISNDSDS PTSPDSIIKS PSKKLMEVSS SSNSSTGPHV 900
901 STAIFGSSLE TCLRLSSHKY QNVYDLPSVV YRCLEYLYKN RGIQEEGIFR LSGSSTVIKT 960
961 LQERFDKEYD VDLCRYNESI EAKDDEASPS LYIGVNTVSG LLKLYLRKLP HLLFGDEQFL1020
1021 SFKRVVDENH NNPVQISLGF KELIESGLVP HANLSLMYAL FELLVRINEN SKFNKMNLRN1080
1081 LCIVFSPTLN IPISMLQPFI TDFACIFQGG EPVKEEEREK VDIHIPQV |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
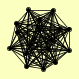
| View Details | Krogan NJ, et al. (2006) |
|
AIM26 APC9 BEM3 FMP27 IRA1 MDM36 MID1 NUP42 REV1 SEC20 SML1 TCO89 TOP1 YDR396W YGL024W ZAP1 ZIM17 |
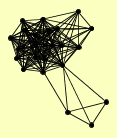
| View Details | Ho Y, et al. (2002) |
|
AVO1 BCK2 BEM3 CCT2 CYS4 DIG1 DIG2 DOP1 GUS1 KSS1 NPA3 PIM1 RNH203 RPA135 RPN6 RVB1 STE11 STE12 STE7 TEC1 TSC11 YIL055C YTA7 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Drees BL, et al. (2001) |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
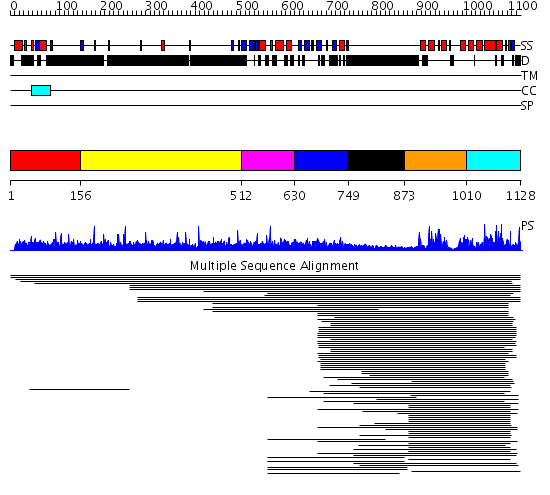
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..155] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [156..511] | 3.030996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [512..629] | 11.309804 | PX domain No confident structure predictions are available. | |
| 4 | View Details | [630..748] | 13.79588 | PH domain No confident structure predictions are available. | |
| 5 | View Details | [749..872] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [873..1009] | 108.383689 | p50 RhoGAP domain | |
| 7 | View Details | [1010..1128] | 108.383689 | p50 RhoGAP domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.87 |
Source: Reynolds et al. (2008)