













| General Information: |
|
| Name(s) found: |
APL6 /
YGR261C
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 809 amino acids |
Gene Ontology: |
|
| Cellular Component: |
AP-3 adaptor complex
[ISS |
| Biological Process: |
vesicle-mediated transport
[TAS Golgi to vacuole transport [IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVDSIHRIAS ALDTAKVITR EAAAVATSKL GESSYTYYSQ NINPQQLVTL LNSRNSREVR 60
61 DAMKRIISIM ASDDDSIDVQ LYFADVVKNI TTNDTKVKRL IHLYLLRFAE NDPNLTLLSI 120
121 NSLQKSLSDS NSELRCFALS ALSDMKMSSL APIILHTVKK LVTDPSAMVR GEVALAIIKL 180
181 YRAGKNDYHE ELLDILKELM ADTDPKVISC AVLAYKECYA DHLELLHGHF RRYCRIIKQL 240
241 DSWSQSYLIE LLIKYCKQYL PKPTVVDKSS EGSPRSCPLP DKYNEIEYPS YEVVNDPDLD 300
301 LFLQSLNCLI YSSNPTVILS CCNALYQLAS PLQMKNTKFI EALVRTVTMT ENQGNKEMLL 360
361 QAIHFLSILD QTLFLPYTKK FYVFPKDPIV ASIWKIQILS TLINESNVKE IFKELKYYVA 420
421 SAHFPENVVI MAVKSLSRCG QLSTSWESHV MKWLIDHMES HNLSASVLDA YVNVIRMLVQ 480
481 KNPTKHLRII FKLADLLTVQ TSLADNARAG IVWLFGEIAS IEFKICPDVL RRLIQNFSNE 540
541 GPETRCQILV LSAKLLSYDI DNFKQAQVTG SEENNQNPPY YDFSGSRISQ MYNAVLYLAK 600
601 YDDEFDIRDR ARMISSLFDS GKYEIVSLLL QAPKPTARSD DFIVSARLET HTPEIKEFFR 660
661 MLPWNTEITE VGETGNDIRE GAELKDYNKY KKSFSSQSFI TNNSARSFTS SSNAKLTGIN 720
721 DGDSNSISGK GNVNTFTSQN GKKYRLQSLD EFFSDIPERK SKPRKIIKVV EESSDEDEDE 780
781 SEESSDDDEY SDSSLGTSSS GTSSSHLEL |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
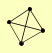
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
APL5 APL6 APM3 APS3 |
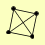
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
APL5 APL6 APM3 APS3 |
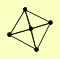
| View Details | Krogan NJ, et al. (2006) |
|
APL5 APL6 APM3 APS3 PUS1 |
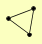
| View Details | Gavin AC, et al. (2006) |
|
APL5 APL6 APM3 |
| View Details | Gavin AC, et al. (2002) |
|
APL1 APL3 APL5 APL6 APM3 APM4 APS2 APS3 CKB1 ENO2 |
| View Details | Qiu et al. (2008) |
|
APL5 APL6 APM3 APS3 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | phosphorylation data - cascade search | Green EM, et al (2005) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
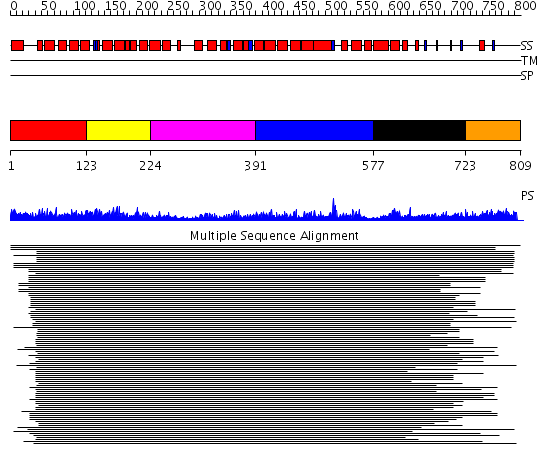
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..122] | 386.228787 | Adaptin beta subunit N-terminal fragment | |
| 2 | View Details | [123..223] | 386.228787 | Adaptin beta subunit N-terminal fragment | |
| 3 | View Details | [224..390] | 386.228787 | Adaptin beta subunit N-terminal fragment | |
| 4 | View Details | [391..576] | 386.228787 | Adaptin beta subunit N-terminal fragment | |
| 5 | View Details | [577..722] | 1.11799 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [723..809] | 11.154902 | Gamma1-adaptin domain |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)