













| General Information: |
|
| Name(s) found: |
LAT1 /
YNL071W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 482 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrial pyruvate dehydrogenase complex
[TAS mitochondrion [IDA |
| Biological Process: |
pyruvate metabolic process
[TAS |
| Molecular Function: |
dihydrolipoyllysine-residue acetyltransferase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSAFVRVVPR ISRSSVLTRS LRLQLRCYAS YPEHTIIGMP ALSPTMTQGN LAAWTKKEGD 60
61 QLSPGEVIAE IETDKAQMDF EFQEDGYLAK ILVPEGTKDI PVNKPIAVYV EDKADVPAFK 120
121 DFKLEDSGSD SKTSTKAQPA EPQAEKKQEA PAEETKTSAP EAKKSDVAAP QGRIFASPLA 180
181 KTIALEKGIS LKDVHGTGPR GRITKADIES YLEKSSKQSS QTSGAAAATP AAATSSTTAG 240
241 SAPSPSSTAS YEDVPISTMR SIIGERLLQS TQGIPSYIVS SKISISKLLK LRQSLNATAN 300
301 DKYKLSINDL LVKAITVAAK RVPDANAYWL PNENVIRKFK NVDVSVAVAT PTGLLTPIVK 360
361 NCEAKGLSQI SNEIKELVKR ARINKLAPEE FQGGTICISN MGMNNAVNMF TSIINPPQST 420
421 ILAIATVERV AVEDAAAENG FSFDNQVTIT GTFDHRTIDG AKGAEFMKEL KTVIENPLEM 480
481 LL |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
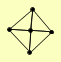
LAT1 LPD1 PDA1 PDB1 PDX1 |
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
LAT1 LPD1 PDA1 PDB1 PDX1 |
| View Details | Krogan NJ, et al. (2006) |

|
LAT1 PDA1 |
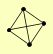
| View Details | Gavin AC, et al. (2006) |
|
LAT1 PDA1 PDB1 PDX1 |
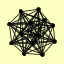
| View Details | Gavin AC, et al. (2002) |
|
APM1 CYM1 DOA4 DUR1,2 HSP42 LAT1 LPD1 PDA1 PDB1 PDX1 PRP46 RPN11 TCM62 |
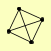
| View Details | Qiu et al. (2008) |
|
LAT1 LPD1 PDA1 PDX1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | De Craene, J., et al. (2006) | |
| View Run | Dam1 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | Dam1 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | sample: hx3 | Hao Xu, et al. (2010) | |
| View Run | Sample: HX11 - both fusion and trans-SNARE complex formation take place. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | Sample: HX13 - trans-SNARE complex forms but fusion is blocked. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX14 - mixture in detergent (control). | Hao Xu, et al. (2010) | |
| View Run | #19 Asynchronous Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #19b Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) | |
| View Run | #19a Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
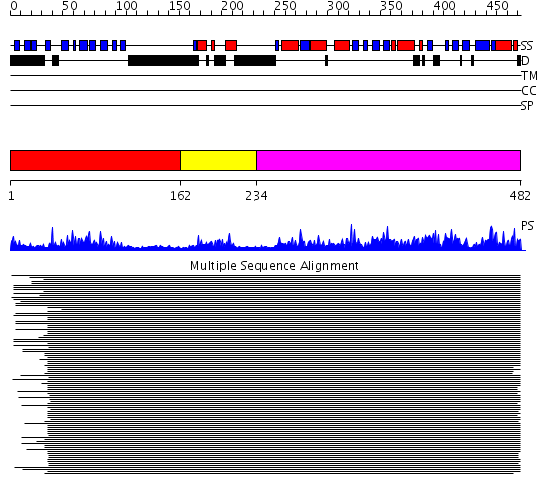
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..161] | 287.025158 | Ipoyl domain of dihydrolipoamide acetyltransferase | |
| 2 | View Details | [162..233] | 28.85005 | E3-binding domain of dihydrolipoamide acetyltransferase | |
| 3 | View Details | [234..482] | 357.08661 | Dihydrolipoamide succinyltransferase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.89 |
Source: Reynolds et al. (2008)