













| General Information: |
|
| Name(s) found: |
ERG6 /
YML008C
[SGD]
|
| Description(s) found:
Found 31 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 383 amino acids |
Gene Ontology: |
|
| Cellular Component: |
lipid particle
[IDA mitochondrion [IDA endoplasmic reticulum [TAS] mitochondrial outer membrane [IDA |
| Biological Process: |
ergosterol biosynthetic process
[TAS]
|
| Molecular Function: |
sterol 24-C-methyltransferase activity
[TAS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSETELRKRQ AQFTRELHGD DIGKKTGLSA LMSKNNSAQK EAVQKYLRNW DGRTDKDAEE 60
61 RRLEDYNEAT HSYYNVVTDF YEYGWGSSFH FSRFYKGESF AASIARHEHY LAYKAGIQRG 120
121 DLVLDVGCGV GGPAREIARF TGCNVIGLNN NDYQIAKAKY YAKKYNLSDQ MDFVKGDFMK 180
181 MDFEENTFDK VYAIEATCHA PKLEGVYSEI YKVLKPGGTF AVYEWVMTDK YDENNPEHRK 240
241 IAYEIELGDG IPKMFHVDVA RKALKNCGFE VLVSEDLADN DDEIPWYYPL TGEWKYVQNL 300
301 ANLATFFRTS YLGRQFTTAM VTVMEKLGLA PEGSKEVTAA LENAAVGLVA GGKSKLFTPM 360
361 MLFVARKPEN AETPSQTSQE ATQ |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
CAF4 ECM27 ERG6 MTF1 PZF1 |
| View Details | Ho Y, et al. (2002) |

|
AAT2 ACH1 ACO1 AFR1 AHP1 ARC1 ARF1 ATP3 CAR2 CDC33 CLU1 COF1 COR1 CPR1 CYR1 CYS4 EGD1 ERG13 ERG6 FPR1 FRS2 GND1 GND2 GRX1 GUS1 HEF3 HHF1, HHF2 LRO1 LSP1 MET6 NTF2 OLA1 OYE2 PFK1 PIL1 PRM2 RNR2 RSN1 SAH1 SCP160 SEC53 SEN15 SES1 SNU13 SOD1 TEF4 THS1 TIF1, TIF2 TMA19 TSA2 UBA1 VMA4 VMA5 WTM1 YNK1 |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
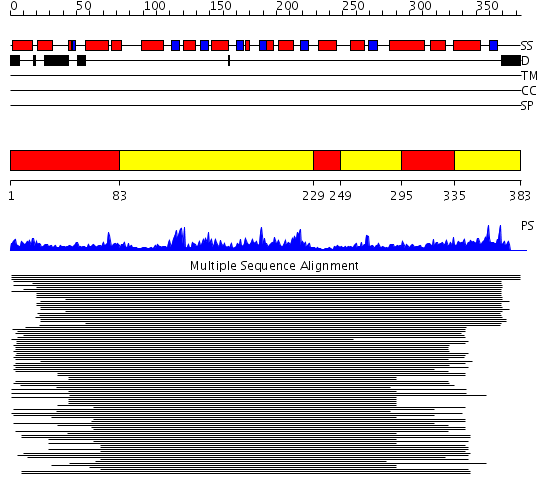
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..82] [229..248] [295..334] |
52.30103 | CmaA1 | |
| 2 | View Details | [83..228] [249..294] [335..383] |
52.30103 | CmaA1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)