













| General Information: |
|
| Name(s) found: |
ERG7 /
YHR072W
[SGD]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 731 amino acids |
Gene Ontology: |
|
| Cellular Component: |
lipid particle
[IDA endoplasmic reticulum [TAS] plasma membrane [TAS |
| Biological Process: |
ergosterol biosynthetic process
[TAS]
|
| Molecular Function: |
lanosterol synthase activity
[TAS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTEFYSDTIG LPKTDPRLWR LRTDELGRES WEYLTPQQAA NDPPSTFTQW LLQDPKFPQP 60
61 HPERNKHSPD FSAFDACHNG ASFFKLLQEP DSGIFPCQYK GPMFMTIGYV AVNYIAGIEI 120
121 PEHERIELIR YIVNTAHPVD GGWGLHSVDK STVFGTVLNY VILRLLGLPK DHPVCAKARS 180
181 TLLRLGGAIG SPHWGKIWLS ALNLYKWEGV NPAPPETWLL PYSLPMHPGR WWVHTRGVYI 240
241 PVSYLSLVKF SCPMTPLLEE LRNEIYTKPF DKINFSKNRN TVCGVDLYYP HSTTLNIANS 300
301 LVVFYEKYLR NRFIYSLSKK KVYDLIKTEL QNTDSLCIAP VNQAFCALVT LIEEGVDSEA 360
361 FQRLQYRFKD ALFHGPQGMT IMGTNGVQTW DCAFAIQYFF VAGLAERPEF YNTIVSAYKF 420
421 LCHAQFDTEC VPGSYRDKRK GAWGFSTKTQ GYTVADCTAE AIKAIIMVKN SPVFSEVHHM 480
481 ISSERLFEGI DVLLNLQNIG SFEYGSFATY EKIKAPLAME TLNPAEVFGD IMVEYPYVEC 540
541 TDSSVLGLTY FHKYFDYRKE EIRTRIRIAI EFIKKSQLPD GSWYGSWGIC FTYAGMFALE 600
601 ALHTVGETYE NSSTVRKGCD FLVSKQMKDG GWGESMKSSE LHSYVDSEKS LVVQTAWALI 660
661 ALLFAEYPNK EVIDRGIDLL KNRQEESGEW KFESVEGVFN HSCAIEYPSY RFLFPIKALG 720
721 MYSRAYETHT L |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Qiu et al. (2008) |

|
ERG7 YEH1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
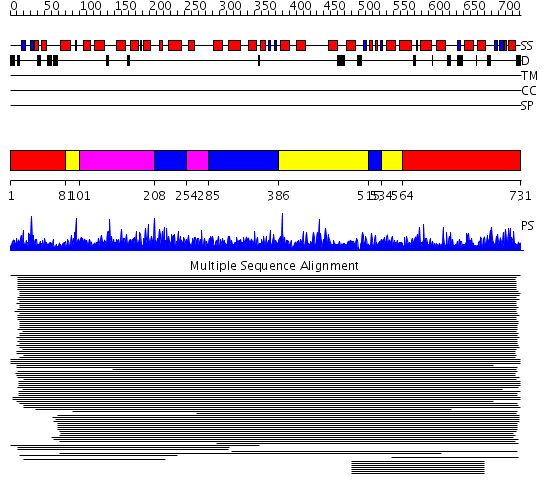
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..80] [564..731] |
377.54902 | Squalene-hopene cyclase | |
| 2 | View Details | [81..100] [386..514] [534..563] |
377.54902 | Squalene-hopene cyclase | |
| 3 | View Details | [101..207] [254..284] |
377.54902 | Squalene-hopene cyclase | |
| 4 | View Details | [208..253] [285..385] [515..533] |
377.54902 | Squalene-hopene cyclase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.89 |
Source: Reynolds et al. (2008)