













| General Information: |
|
| Name(s) found: |
NUP120 /
YKL057C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1037 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear pore
[IDA |
| Biological Process: |
mRNA export from nucleus
[IMP rRNA export from nucleus [TAS nuclear pore organization [IMP snRNP protein import into nucleus [TAS mRNA-binding (hnRNP) protein import into nucleus [TAS snRNA export from nucleus [TAS chromosome localization [IMP tRNA export from nucleus [TAS NLS-bearing substrate import into nucleus [TAS DNA metabolic process [IGI protein export from nucleus [TAS ribosomal protein import into nucleus [TAS |
| Molecular Function: |
structural molecule activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MACLSRIDAN LLQYYEKPEP NNTVDLYVSN NSNNNGLKEG DKSISTPVPQ PYGSEYSNCL 60
61 LLSNSEYICY HFSSRSTLLT FYPLSDAYHG KTINIHLPNA SMNQRYTLTI QEVEQQLLVN 120
121 VILKDGSFLT LQLPLSFLFS SANTLNGEWF HLQNPYDFTV RVPHFLFYVS PQFSVVFLED 180
181 GGLLGLKKVD GVHYEPLLFN DNSYLKSLTR FFSRSSKSDY DSVISCKLFH ERYLIVLTQN 240
241 CHLKIWDLTS FTLIQDYDMV SQSDSDPSHF RKVEAVGEYL SLYNNTLVTL LPLENGLFQM 300
301 GTLLVDSSGI LTYTFQNNIP TNLSASAIWS IVDLVLTRPL ELNVEASYLN LIVLWKSGTA 360
361 SKLQILNVND ESFKNYEWIE SVNKSLVDLQ SEHDLDIVTK TGDVERGFCN LKSRYGTQIF 420
421 ERAQQILSEN KIIMAHNEDE EYLANLETIL RDVKTAFNEA SSITLYGDEI ILVNCFQPYN 480
481 HSLYKLNTTV ENWFYNMHSE TDGSELFKYL RTLNGFASTL SNDVLRSISK KFLDIITGEL 540
541 PDSMTTVEKF TDIFKNCLEN QFEITNLKIL FDELNSFDIP VVLNDLINNQ MKPGIFWKKD 600
601 FISAIKFDGF TSIISLESLH QLLSIHYRIT LQVLLTFVLF DLDTEIFGQH ISTLLDLHYK 660
661 QFLLLNLYRQ DKCLLAEVLL KDSSEFSFGV KFFNYGQLIA YIDSLNSNVY NASITENSFF 720
721 MTFFRSYIIE NTSHKNIRFF LENVECPFYL RHNEVQEFMF AMTLFSCGNF DQSYEIFQLH 780
781 DYPEAINDKL PTFLEDLKSE NYHGDSIWKD LLCTFTVPYR HSAFYYQLSL LFDRNNSQEF 840
841 ALKCISKSAE YSLKEIQIEE LQDFKEKQHI HYLNLLIHFR MFEEVLDVLR LGHECLSDTV 900
901 RTNFLQLLLQ EDIYSRDFFS TLLRLCNAHS DNGELYLRTV DIKIVDSILS QNLRSGDWEC 960
961 FKKLYCFRML NKSERAAAEV LYQYILMQAD LDVIRKRKCY LMVINVLSSF DSAYDQWILN1020
1021 GSKVVTLTDL RDELRGL |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
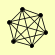
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
NUP120 NUP145 NUP84 NUP85 SEC13 SEH1 |
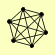
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
NUP120 NUP145 NUP84 NUP85 SEC13 SEH1 |
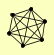
| View Details | Krogan NJ, et al. (2006) |
|
MEX67 NUP120 NUP145 NUP84 NUP85 SEH1 |
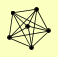
| View Details | Gavin AC, et al. (2006) |
|
NUP120 NUP145 NUP84 NUP85 SEC13 SEH1 |
| View Details | Gavin AC, et al. (2002) |
|
CKA1 CKA2 CKB1 DBP2 MRPS28 NUP120 NUP145 NUP84 NUP85 RPP2B SEC13 SEC16 SEC23 SEC31 SEH1 STM1 |
| View Details | Ho Y, et al. (2002) |
|
NUP120 NUP84 PHO86 |
| View Details | Qiu et al. (2008) |
|
ASM4 KAP123 NIC96 NUP100 NUP116 NUP120 NUP133 NUP145 NUP157 NUP159 NUP170 NUP188 NUP57 NUP82 NUP84 NUP85 POM152 SEC13 SEH1 SUS1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
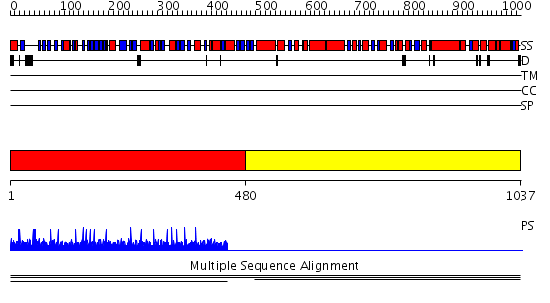
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..479] | 1.002 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [480..1037] | 1.002001 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [590..745] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [746..1037] | 0.975 | Vesicular transport protein sec17 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 |
|
||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)