













| General Information: |
|
| Name(s) found: |
YPR097W
[SGD]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1073 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
phosphoinositide binding
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MITQDTPALN PTEEHYLKRE LLRCQLDYEI GKLNDQFALR KFGYPFSPND PTAPQPISNN 60
61 DSSPVLGGKG HFSVNYPMLS YVLQEFISTF PLLSTNLLVD EKFWQSKVQV FFEHFMSLGF 120
121 SESYDREEAS KRKKVSKKLS KVILLLFNSG VGSFQEQAYY NEDKFVLQSG QARKRSNIEK 180
181 FAMPTRENLE NLLTNESVFI NGWDVNIISV FNKNSRKCTE SVDNDKSSKS TPTSSPKSHA 240
241 IKSFASTSKW MKNAFNNTIN STINSMPESS ASLFSKLSLG VPSTKSKQSR KHHYFLIKIK 300
301 KQDDDDQDNS NEENSNLDHH AGYFYVTRTY SDFKKLSHDL KSEFPGKKCP RLPHRNKKVT 360
361 SMITKTEVLH NGQTKSAARE KIVNTFDTDL QSASESDNSS FLQTTNELSA TETVLTEKET 420
421 ETLRKNILNE IKEEDNIDED EYEEEGEGEE SDFDEYKDAS DSKINTLVGE KMRTSLRQYL 480
481 RTLCKDAEVS QSSSIRRFFL SGPNLDIKDI NPKIADDIRN RALIDVSNLE NQIRFQQMAL 540
541 EKSLKLQDSM KDFKTSLLKD EKYLMSLLVE IKDNTKVEDL SPLLQDFVEW CKIYISSMIY 600
601 QMFLGNDNSY ELYTQIRRLH KLMPYTVMGQ IMKFTNPIAI MRGMIELFMA QPFGGHSLLQ 660
661 TMFSTILTDD LKTQKVAIKE LERKIAEMDP GASVVTKCLK DFVFNNDTKD EHDTKLFTMD 720
721 AVNAESESMN MPVPLIVLMK SAAANLIPDE VVAGLIESYS SWKLQKEDTD ALNVTSEDQS 780
781 GIYFTHVKDL WQLYIKEHDK QLMRQLWQDP ELTQMLKAIV TMIYEPMVKI FKVARMDVAL 840
841 KNFEKFMGDL IRLVDDVING QLGVSTQFDV VEEIHNLVTK HQDAFFEFIH DVYLNDSEGI 900
901 FEGFITWITT IVKFLQKSKF GGPSERIDFN KLICRDDIDI DVKLLKVQVN NVLNKKIGAR 960
961 KIYKKLLDLK VKQGTKQNNK HAAGILQKNW SDINSLVMPS SSGSFGLGDG DLVDLDLDTG1020
1021 DYDFLHKENE VELEKQYKDL LNLVVDESEI DKLRSQVFAQ ELKNYLEAQI AKK |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
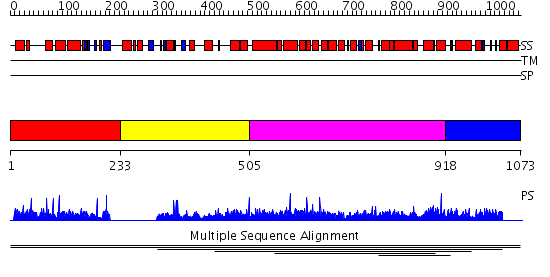
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..232] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [233..504] | 20.60206 | PX domain No confident structure predictions are available. | |
| 3 | View Details | [505..917] | 1.004919 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [918..1073] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [563..771] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [772..1073] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||
| 1 | No functions predicted. | ||||||||||||
| 2 | No functions predicted. | ||||||||||||
| 3 |
|
||||||||||||
| 4 | No functions predicted. | ||||||||||||
| 5 | No functions predicted. | ||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)