













| General Information: |
|
| Name(s) found: |
CCM1 /
YGR150C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 864 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MYMARCGPKN NVLCFPFQLS FLFSKRLINK RFKYTLQTED EKNMMGSLSK NKIITPEDVE 60
61 FKLAQLREFS NTLKERIHNT KSVNSDGHQS NSIAPISEDS RNVNVTKTSS VPNEEKSKNL 120
121 SDLIHSSFLE KMDHLVPKVI RERVADDDIL AKNLFDRSHS NWAPVIDRLY VSEKRFMDID 180
181 SREFSVWLNG TVKYLPFHSI LHLDEMLLEQ INGDVVKFNT HMYECIFNNL GNLKPTNFNQ 240
241 DGTNDKVILK MKELLERYDK ALKITEERIN KKEGFPSKVP KMTQAILNNC LKYSTKCSSF 300
301 HDMDYFITKF RDDYGITPNK QNLTTVIQFY SRKEMTKQAW NTFDTMKFLS TKHFPDICTY 360
361 NTMLRICEKE RNFPKALDLF QEIQDHNIKP TTNTYIMMAR VLASSSSNAV VSEGKSDSLR 420
421 LLGWKYLHEL EDKNLYRHKK DDLNLFLAMM ALAAFDGDIE LSRALYYLFI AKKYKTLCAN 480
481 WKGNILVDQD TIWKSTLMPE MLNYLMLAYA RFDPRNLPVL SGYEKGIELR RKFLREFDSS 540
541 MRLDDTDKLV KFKLPFLPIS DLNSEAQVLA ESNAIWSFNM ENGGTRNTLT SSNEAALEDI 600
601 KKYRQLLDSF AQEAEDFNEF KFKVMYEVTK MQRESINVNV FNKISLHTYL SIPINLKQQK 660
661 EFLRRLTFFT FQQHEFEAVI KRLYEGYRNI PSSHTRDQNS ISTEAISVSK PETTEDLNLI 720
721 MHDIWYITCL RHKIMMDTTL YELVMKAAIE FQNEDLAKKV WNDRGKFRTT VPFLKMDQRI 780
781 RIAKDQKFAH LMVEFFTKQG KYSDAIAIIL SSKNRFNWTY SMVRNLHKAL EEIEDRNSVE 840
841 ILLDVVNKKS HAKALKWEEQ ELNM |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
CCM1 MRP17 |
| View Details | Gavin AC, et al. (2002) |

|
BEM2 CCM1 ECM16 EHD3 FYV4 GEP3 HSC82 MRP1 MRP10 MRP13 MRP17 MRP21 MRP4 MRP51 MRPS16 MRPS17 MRPS18 MRPS28 MRPS5 MRPS8 MRPS9 NAM9 PAP2 PET123 PSD2 PUS7 RIX1 RSM10 RSM19 RSM22 RSM23 RSM24 RSM25 RSM26 RSM27 RSM7 TIF4631 UBP10 UTP20 UTP22 |
| View Details | Ho Y, et al. (2002) |

|
CBF5 CBP2 CCM1 CDC33 CIC1 DBP7 ENP2 GBP2 HAS1 HMO1 HTB2 IMD1 IMD3 ISA1 KRE33 KRI1 MGM101 MSS116 NOC2 NOP12 NOP6 NPL3 PET127 POL5 PRP43 PUF6 PWP1 RLI1 RRP12 RRP5 TIF4631 TRA1 TSR1 URB1 URB2 UTP10 UTP20 YPP1 YRA1 YTM1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
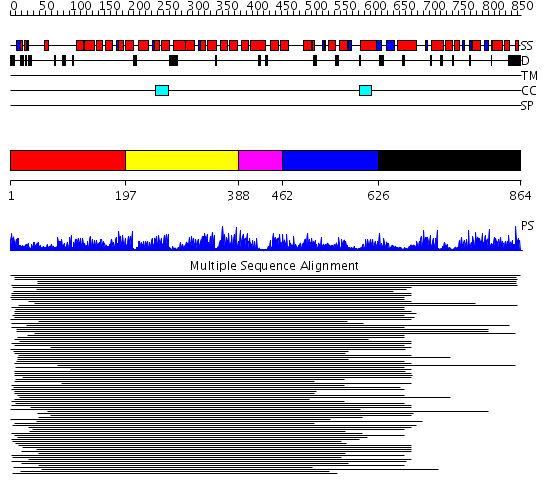
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..196] | 14.38 | Peroxin pex5 (peroxisomal targeting signal 1 (PTS1) receptor) | |
| 2 | View Details | [197..387] | 12.12 | Constant regulatory domain of protein phosphatase 2a, pr65alpha | |
| 3 | View Details | [388..461] | 12.12 | Constant regulatory domain of protein phosphatase 2a, pr65alpha | |
| 4 | View Details | [462..625] | 12.12 | Constant regulatory domain of protein phosphatase 2a, pr65alpha | |
| 5 | View Details | [626..864] | 12.12 | Constant regulatory domain of protein phosphatase 2a, pr65alpha |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. | |||||||||
| 4 | No functions predicted. | |||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)