













| General Information: |
|
| Name(s) found: |
RGD2 /
YFL047W
[SGD]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 714 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA cellular bud neck [IDA |
| Biological Process: |
small GTPase mediated signal transduction
[IC |
| Molecular Function: |
Rho GTPase activator activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLSFCDYFWS EDLVSGLDVL FDRLYHGCEQ CDLFIQLFAS RMQFEVSHGR QLFGIEAGMD 60
61 NLKAVQEDED EGVTVSRALR GILQEMSQEG THHLTIASNI ESLVLQPFSK WCIEHRERIQ 120
121 YSEKTLLTNV NNFRKSKKYV GKLEKEYFNK CRQLEEFKRT HFNEDELANA MKSLKIQNKY 180
181 EEDVAREKDH RFFNRIAGID FDYKTMKETL QLLLTKLPKT DYKLPLISYS LSNTNNGGEI 240
241 TKFLLDHMSL KDIDQAETFG QDLLNLGFLK YCNGVGNTFV NSKKFQYQWK NTAYMFANVP 300
301 MPGSEEPTTG ESLISRFNNW DGSSAKEIIQ SKIGNDQGAA KIQAPHISDN ERTLFRMMDA 360
361 LAASDKKYYQ ECFKMDALRC SVEELLIDHL SFMEKCESDR LNAIKKATLD FCSTLGNKIS 420
421 SLRLCIDKML TLENDIDPTA DLLQLLVKYK TGSFKPQAIV YNNYYNPGSF QNFGVDLETR 480
481 CRLDKKVVPL IISSIFSYMD KIYPDLPNDK VRTSIWTDSV KLSLTHQLRN LLNKQQFHNE 540
541 GEIFDILSTS KLEPSTIASV VKIYLLELPD PLIPNDVSDI LRVLYLDYPP LVETALQNST 600
601 SSPENQQDDD NEEGFDTKRI RGLYTTLSSL SKPHIATLDA ITTHFYRLIK ILKMGENGNE 660
661 VADEFTVSIS QEFANCIIQS KITDDNEIGF KIFYDLLTHK KQIFHELKRQ NSKN |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Qiu et al. (2008) |
|
LRG1 RGD2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #13 Mitotic Prep2-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
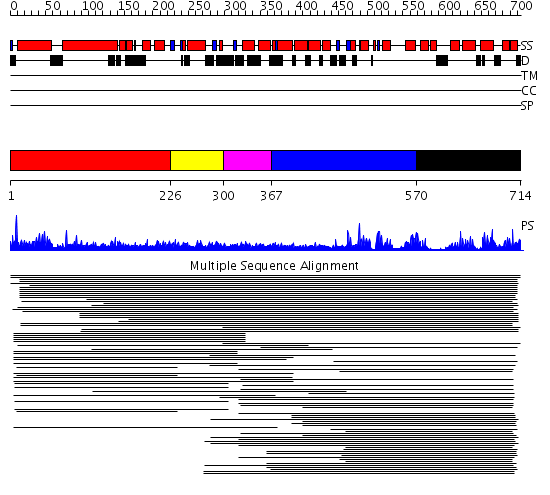
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..225] | 9.07 | Apolipoprotein A-I | |
| 2 | View Details | [226..299] | 12.180456 | Domain found in Dishevelled, Egl-10, and Pleckstrin No confident structure predictions are available. | |
| 3 | View Details | [300..366] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [367..569] | 38.0 | p50 RhoGAP domain | |
| 5 | View Details | [570..714] | 38.0 | p50 RhoGAP domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)