













| General Information: |
|
| Name(s) found: |
LRG1 /
YDL240W
[SGD]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1017 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA cellular bud neck [IDA mitochondrion [IDA |
| Biological Process: |
fungal-type cell wall biogenesis
[IGI small GTPase mediated signal transduction [IC |
| Molecular Function: |
Rho GTPase activator activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MIQNSAGYRS LNTASPMTVQ VKNQKKICAR CNKLVIPDSQ RTKTTLKALG KYYHESCFTC 60
61 QDCQKPLKPK YFPYQVDKTS ESILLCQYDY FRRHNLLCHV CDTPLRGLYY TAFGYRYDEE 120
121 HFSCTICATP CGVKKCFMYG NQLYCKYHFL KYFSKRCKGC EFPISDQYIE FPKGEEIHCW 180
181 HPECYGIHKY WHVNLAAETV GLQYLPKLEY NPNSGDKDIN PTAYELDKQM QAFNFILSKT 240
241 WSVLYRFEEE AASCISDMFQ YLTSNDQLKG IESTGLLVLK IDCLFRGLDT LNLSTNKSMP 300
301 VNSDQECIEN NAMAASKYSK FPKNLSTKIM IYLQLLRKLG TENKNETITI SSFMSVITGL 360
361 AHFLKLLTRF GLYTALENNK LTHSVNPLLR FLREVEKNEL FENNPFQYIK TPVNATDSCA 420
421 GCNKYIQEEC IQFYEHRWHI ACFTCSSCHK NINPRSLTDP TFNKEKKKIL CSHCSIDDPA 480
481 SVPGFKFVTK LAQLIFLLKI ALVKSRTVML KSKASNKVGR NSLQSTMLKE HTYIRTLNDI 540
541 KRLRSRRESV RVTHNKQQAR KSVILETAET DLNDPTKQGD SKNLVIQTDD PSSSQQVSTR 600
601 ENVFSNTKTL TLDDISRIVA AEQARELRPN AFAHFKKLKE TDDETSNVVP KKSGVYYSEL 660
661 STMELSMIRA ISLSLLAGKQ LISKTDPNYT SLVSMVFSNE KQVTGSFWNR MKIMMSMEPK 720
721 KPITKTVFGA PLDVLCEKWG VDSDLGVGPV KIRIPIIIDE LISSLRQMDM SVEGIFRKNG 780
781 NIRRLRELTA NIDSNPTEAP DFSKENAIQL SALLKKFIRE LPQPILSTDL YELWIKAAKI 840
841 DLEDEKQRVI LLIYSLLPTY NRNLLEALLS FLHWTSSFSY IENEMGSKMD IHNLSTVITP 900
901 NILYLRHKEI SNDNVPDEPE SGLVDSFAQN KGENYFLAIE IVDYLITHNE EMAMVPKFLM 960
961 NLLKDVQLQK LDNYESINHF ISTVMQSKTI DYSECDIKTP VTVKDSTTTV IQGEINK |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Qiu et al. (2008) |
|
BUD5 CDC24 LRG1 RGD1 RGD2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) | |
| View Run | #13 Mitotic Prep2-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
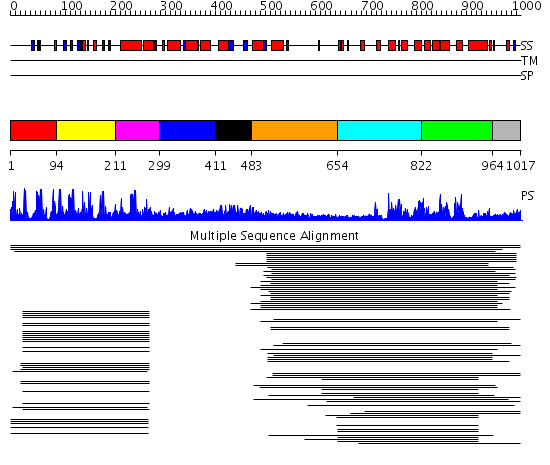
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..93] | 10.30103 | Cysteine-rich (intestinal) protein, CRP, CRIP | |
| 2 | View Details | [94..210] | 10.30103 | Cysteine-rich (intestinal) protein, CRP, CRIP | |
| 3 | View Details | [211..298] | 4.127997 | View MSA. Confident ab initio structure predictions are available. | |
| 4 | View Details | [299..410] | N/A | Confident ab initio structure predictions are available. | |
| 5 | View Details | [411..482] | 15.431798 | LIM domain No confident structure predictions are available. | |
| 6 | View Details | [483..653] | 1.106979 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [654..821] | 207.927757 | p50 RhoGAP domain | |
| 8 | View Details | [822..963] | 207.927757 | p50 RhoGAP domain | |
| 9 | View Details | [964..1017] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)