













| General Information: |
|
| Name(s) found: |
TRZ1 /
YKR079C
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 838 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA mitochondrion [IDA |
| Biological Process: |
tRNA 3'-trailer cleavage
[IDA |
| Molecular Function: |
purine nucleotide binding
[ISS 3'-tRNA processing endoribonuclease activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFTFIPITHP TSDTKHPLLL VQSAHGEKYF FGKIGEGSQR SLTENKIRIS KLKDIFLTGE 60
61 LNWSDIGGLP GMILTIADQG KSNLVLHYGN DILNYIVSTW RYFVFRFGID LNDHIMKDKE 120
121 VYKDKIIAVK SFNVLKNGGE DRLGVFDSFQ KGVLRSIVAK MFPKHAPTDR YDPSSDPHLN 180
181 VELPDLDAKV EVSTNYEISF SPVRGKFKVE EAIKLGVPKG PLFAKLTKGQ TITLDNGIVV 240
241 TPEQVLENER HFAKVLILDI PDDLYLNAFV EKFKDYDCAE LGMVYYFLGD EVTINDNLFA 300
301 FIDIFEKNNY GKVNHMISHN KISPNTISFF GSALTTLKLK ALQVNNYNLP KTDRVFSKDF 360
361 YDRFDTPLSR GTSMCKSQEE PLNTIIEKDN IHIFSQNKTV TFEPFRMNEE PMKCNINGEV 420
421 ADFSWQEIFE EHVKPLEFPL ADVDTVINNQ LHVDNFNNSA EKKKHVEIIT LGTGSALPSK 480
481 YRNVVSTLVK VPFTDADGNT INRNIMLDAG ENTLGTIHRM FSQLAVKSIF QDLKMIYLSH 540
541 LHADHHLGII SVLNEWYKYN KDDETSYIYV VTPWQYHKFV NEWLVLENKE ILKRIKYISC 600
601 EHFINDSFVR MQTQSVPLAE FNEILKENSN QESNRKLELD RDSSYRDVDL IRQMYEDLSI 660
661 EYFQTCRAIH CDWAYSNSIT FRMDENNEHN TFKVSYSGDT RPNIEKFSLE IGYNSDLLIH 720
721 EATLENQLLE DAVKKKHCTI NEAIGVSNKM NARKLILTHF SQRYPKLPQL DNNIDVMARE 780
781 FCFAFDSMIV DYEKIGEQQR IFPLLNKAFV EEKEEEEDVD DVESVQDLEV KLKKHKKN |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
NUC1 TRZ1 |
| View Details | Hazbun TR, et al. (2003) |

|
NUC1 TRZ1 YMR099C |
| View Details | Gavin AC, et al. (2006) |
|
NUC1 TRZ1 YMR099C |
| View Details | Gavin AC, et al. (2002) |

|
NUC1 TRZ1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Hazbun TR, et al. (2003) |
| View Data |
|
Hazbun TR, et al. (2003) |
| View Data |
|
Hazbun TR, et al. (2003) |
| View Data |
|
Hazbun TR, et al. (2003) |
| View Data |
|
Unpublished Data |
| View Data |
|
Unpublished Data |
| View Data |
|
Huh WK, et al. (2003) |
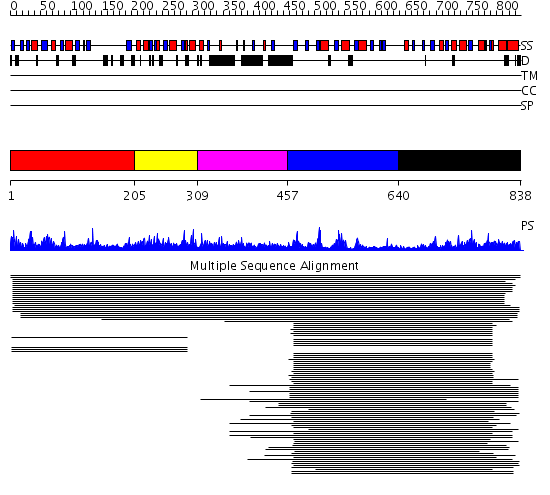
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..204] | 1.66 | Glyoxalase II (hydroxyacylglutathione hydrolase) | |
| 2 | View Details | [205..308] | 1.66 | Glyoxalase II (hydroxyacylglutathione hydrolase) | |
| 3 | View Details | [309..456] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [457..639] | 4.30103 | Rubredoxin oxygen:oxidoreductase (ROO), C-terminal domain; Rubredoxin oxygen:oxidoreductase (ROO), N-terminal domain | |
| 5 | View Details | [640..838] | 3.267975 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||
| 4 |
|
|||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)