













| General Information: |
|
| Name(s) found: |
YLL007C
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 665 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKHNRQMPDE SLENIKVLLN PKLGKPVKSL TSAQSKACYH TLISNKNLNK TSDEYEKLLA 60
61 NYILLCDEKY LCKTVIPDSR FWAILCDNCQ KLRSETLVAN LIRIFNVALK CQDSNKNEVI 120
121 VSICHISREN SQLIGILLQL LSQRPIHIPL FTDTILCITL FLKCSLTLCE TSLSHAVEFV 180
181 PRILILLFQY NFPASMSELL YIEDLQPLIL EEFVPLKQRL INFLSSVSID DYSCSLKADL 240
241 LTAIKDNSVF QKGLEMEMGD LPSINLLNAY DTFTFLNSPN GSFKRLYTEQ LLFGENDFPL 300
301 YEAIFKLSDQ FRRLFNLSGK KENQYSDSER DLKLQIATAV LNRQTCFYKT LELFLRFWIE 360
361 SLAKSQSDLV SLLNLAIITL KYVCLSSSDL EAAIQTKSLL KTQVVALDSM RYKFARTLQL 420
421 DSIKKEQYRT WSSSIASFDT MLSGQVRDYV RHQRLLQLQK GTWVYAENPL NPEAGTPKVY 480
481 FLIVSDNHAN LLAREFETQT NDLPYLFDNK ILTSPGSEAL ANGRTKVVVL KHITSFKSIE 540
541 LTTPSRRTSS NVYIKLDEAN VYTGVELKDR NDRTVLKFYL DTEEGRYIWL DGLKLISPFQ 600
601 HEDISEDTKE QIDTLFDLRK NVQMINLNVR QDIIVPPPEP SDEDEDEEFY NLETLKKVTQ 660
661 NFYFD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Hazbun TR, et al. (2003) | ||
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
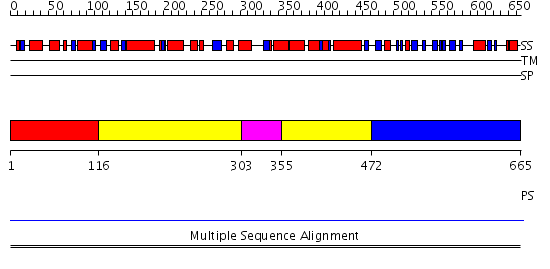
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..115] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [116..302] [355..471] |
7.59 | Hemolysin E (HlyE, ClyA, SheA) | |
| 3 | View Details | [303..354] | 7.59 | Hemolysin E (HlyE, ClyA, SheA) | |
| 4 | View Details | [472..665] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. | |||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)